
Vibrio azureus NBRC 104587
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio; Vibrio harveyi group; Vibrio azureus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
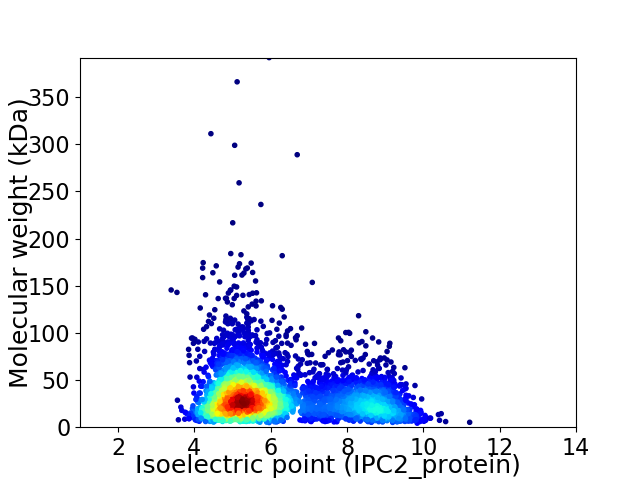
Virtual 2D-PAGE plot for 4147 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U3A2C3|U3A2C3_9VIBR DNA topoisomerase 3 OS=Vibrio azureus NBRC 104587 OX=1219077 GN=topB PE=3 SV=1
MM1 pKa = 7.38IAKK4 pKa = 8.17TKK6 pKa = 10.23LASLCLLSLASANIYY21 pKa = 10.16ASSVIDD27 pKa = 4.07LDD29 pKa = 4.22FSNHH33 pKa = 5.86LEE35 pKa = 4.05ATNSSVSWAGPVFTGPDD52 pKa = 3.18MHH54 pKa = 6.47FVGVGTHH61 pKa = 6.83DD62 pKa = 4.17GKK64 pKa = 9.61TIDD67 pKa = 3.85AKK69 pKa = 9.87VTSSVFGDD77 pKa = 3.84ATFLWHH83 pKa = 6.99APNYY87 pKa = 8.64NQLSGSQPLGDD98 pKa = 4.49IGFLYY103 pKa = 8.56QTNSPGNAGLTYY115 pKa = 10.63VFEE118 pKa = 4.9FFDD121 pKa = 4.07GTDD124 pKa = 3.54GLSGTFSVPYY134 pKa = 9.8IIPEE138 pKa = 4.05FEE140 pKa = 4.28FIGYY144 pKa = 9.75DD145 pKa = 3.11IDD147 pKa = 4.14GEE149 pKa = 4.46PVQSEE154 pKa = 3.95QVRR157 pKa = 11.84VFKK160 pKa = 11.31SEE162 pKa = 3.71GFYY165 pKa = 10.55SYY167 pKa = 11.3QLGSGAASLTAQEE180 pKa = 4.76SPDD183 pKa = 3.64GQSVLFTGPGTNFSEE198 pKa = 4.61TDD200 pKa = 2.76TSGAVKK206 pKa = 8.87FTYY209 pKa = 10.46KK210 pKa = 9.25NTSIVTLQFEE220 pKa = 4.87TVTTSASGFPNPIFSAFDD238 pKa = 3.93GNWDD242 pKa = 3.59LAGFDD247 pKa = 4.25TPTTSNSDD255 pKa = 3.14FDD257 pKa = 4.93YY258 pKa = 11.53GDD260 pKa = 4.2APDD263 pKa = 3.63TYY265 pKa = 9.74KK266 pKa = 10.63TLLASDD272 pKa = 5.41GPQHH276 pKa = 6.61ALSSSLYY283 pKa = 10.17LGSSVDD289 pKa = 4.23ADD291 pKa = 4.02PDD293 pKa = 3.87GQPNPTSTGDD303 pKa = 3.81DD304 pKa = 3.4FDD306 pKa = 5.72VDD308 pKa = 4.3GNDD311 pKa = 4.66DD312 pKa = 4.78DD313 pKa = 6.73GITPLTNFEE322 pKa = 4.5IGLDD326 pKa = 3.63ALINAQVVGEE336 pKa = 4.77GYY338 pKa = 10.33LHH340 pKa = 7.38AWADD344 pKa = 2.95WDD346 pKa = 3.95MNGTFDD352 pKa = 5.6ADD354 pKa = 3.61EE355 pKa = 4.79QILKK359 pKa = 10.25NYY361 pKa = 10.33NVTTGANVIPIRR373 pKa = 11.84VGDD376 pKa = 3.84DD377 pKa = 3.77AIVGNVQTRR386 pKa = 11.84FRR388 pKa = 11.84LSSSPNLPSTGYY400 pKa = 10.86AGDD403 pKa = 4.07GEE405 pKa = 4.57VEE407 pKa = 4.01DD408 pKa = 4.75RR409 pKa = 11.84VFDD412 pKa = 3.95VVDD415 pKa = 4.06PEE417 pKa = 4.51TTIQTSDD424 pKa = 3.82YY425 pKa = 8.3YY426 pKa = 10.71TAAFEE431 pKa = 5.39DD432 pKa = 3.98NWPEE436 pKa = 4.15MGDD439 pKa = 3.36FDD441 pKa = 6.53LNDD444 pKa = 2.91VVTYY448 pKa = 10.65YY449 pKa = 9.03RR450 pKa = 11.84TKK452 pKa = 10.78VISKK456 pKa = 9.88DD457 pKa = 3.63GNVLRR462 pKa = 11.84LDD464 pKa = 3.37IEE466 pKa = 4.57GTITAYY472 pKa = 10.08GASYY476 pKa = 11.43SNGLAWKK483 pKa = 9.92LDD485 pKa = 3.6GFAEE489 pKa = 4.47SDD491 pKa = 3.76VNLQLSRR498 pKa = 11.84VTKK501 pKa = 10.62NGQLRR506 pKa = 11.84NGISPFTGEE515 pKa = 4.62DD516 pKa = 3.16KK517 pKa = 11.11SVASPGGDD525 pKa = 2.84LVVVASVNLLDD536 pKa = 5.98DD537 pKa = 3.83IPIHH541 pKa = 5.96GEE543 pKa = 4.1CIYY546 pKa = 10.98HH547 pKa = 5.93RR548 pKa = 11.84TNPSCNPSLEE558 pKa = 4.19SSQMTFSVSLPFNDD572 pKa = 3.95GSEE575 pKa = 4.18PTVTSLLPLDD585 pKa = 3.85GHH587 pKa = 7.01DD588 pKa = 3.87PFIFASGEE596 pKa = 3.95GSYY599 pKa = 10.25HH600 pKa = 6.99GEE602 pKa = 3.98IFTVPPGKK610 pKa = 10.14DD611 pKa = 3.62LEE613 pKa = 4.21IHH615 pKa = 5.98TADD618 pKa = 4.54FPPTSRR624 pKa = 11.84GTLVSTFYY632 pKa = 10.79EE633 pKa = 4.28QADD636 pKa = 3.87DD637 pKa = 4.81RR638 pKa = 11.84SDD640 pKa = 3.59PSISKK645 pKa = 10.4YY646 pKa = 10.65YY647 pKa = 9.36RR648 pKa = 11.84TDD650 pKa = 3.2GNLPWGIIIPSIWNHH665 pKa = 5.04PSEE668 pKa = 4.83YY669 pKa = 9.31IDD671 pKa = 3.95VSVAYY676 pKa = 9.5PDD678 pKa = 3.6FAEE681 pKa = 4.3WAVSGGSSKK690 pKa = 9.13PTWYY694 pKa = 10.66LNPEE698 pKa = 4.28SSQTWTTSDD707 pKa = 2.96
MM1 pKa = 7.38IAKK4 pKa = 8.17TKK6 pKa = 10.23LASLCLLSLASANIYY21 pKa = 10.16ASSVIDD27 pKa = 4.07LDD29 pKa = 4.22FSNHH33 pKa = 5.86LEE35 pKa = 4.05ATNSSVSWAGPVFTGPDD52 pKa = 3.18MHH54 pKa = 6.47FVGVGTHH61 pKa = 6.83DD62 pKa = 4.17GKK64 pKa = 9.61TIDD67 pKa = 3.85AKK69 pKa = 9.87VTSSVFGDD77 pKa = 3.84ATFLWHH83 pKa = 6.99APNYY87 pKa = 8.64NQLSGSQPLGDD98 pKa = 4.49IGFLYY103 pKa = 8.56QTNSPGNAGLTYY115 pKa = 10.63VFEE118 pKa = 4.9FFDD121 pKa = 4.07GTDD124 pKa = 3.54GLSGTFSVPYY134 pKa = 9.8IIPEE138 pKa = 4.05FEE140 pKa = 4.28FIGYY144 pKa = 9.75DD145 pKa = 3.11IDD147 pKa = 4.14GEE149 pKa = 4.46PVQSEE154 pKa = 3.95QVRR157 pKa = 11.84VFKK160 pKa = 11.31SEE162 pKa = 3.71GFYY165 pKa = 10.55SYY167 pKa = 11.3QLGSGAASLTAQEE180 pKa = 4.76SPDD183 pKa = 3.64GQSVLFTGPGTNFSEE198 pKa = 4.61TDD200 pKa = 2.76TSGAVKK206 pKa = 8.87FTYY209 pKa = 10.46KK210 pKa = 9.25NTSIVTLQFEE220 pKa = 4.87TVTTSASGFPNPIFSAFDD238 pKa = 3.93GNWDD242 pKa = 3.59LAGFDD247 pKa = 4.25TPTTSNSDD255 pKa = 3.14FDD257 pKa = 4.93YY258 pKa = 11.53GDD260 pKa = 4.2APDD263 pKa = 3.63TYY265 pKa = 9.74KK266 pKa = 10.63TLLASDD272 pKa = 5.41GPQHH276 pKa = 6.61ALSSSLYY283 pKa = 10.17LGSSVDD289 pKa = 4.23ADD291 pKa = 4.02PDD293 pKa = 3.87GQPNPTSTGDD303 pKa = 3.81DD304 pKa = 3.4FDD306 pKa = 5.72VDD308 pKa = 4.3GNDD311 pKa = 4.66DD312 pKa = 4.78DD313 pKa = 6.73GITPLTNFEE322 pKa = 4.5IGLDD326 pKa = 3.63ALINAQVVGEE336 pKa = 4.77GYY338 pKa = 10.33LHH340 pKa = 7.38AWADD344 pKa = 2.95WDD346 pKa = 3.95MNGTFDD352 pKa = 5.6ADD354 pKa = 3.61EE355 pKa = 4.79QILKK359 pKa = 10.25NYY361 pKa = 10.33NVTTGANVIPIRR373 pKa = 11.84VGDD376 pKa = 3.84DD377 pKa = 3.77AIVGNVQTRR386 pKa = 11.84FRR388 pKa = 11.84LSSSPNLPSTGYY400 pKa = 10.86AGDD403 pKa = 4.07GEE405 pKa = 4.57VEE407 pKa = 4.01DD408 pKa = 4.75RR409 pKa = 11.84VFDD412 pKa = 3.95VVDD415 pKa = 4.06PEE417 pKa = 4.51TTIQTSDD424 pKa = 3.82YY425 pKa = 8.3YY426 pKa = 10.71TAAFEE431 pKa = 5.39DD432 pKa = 3.98NWPEE436 pKa = 4.15MGDD439 pKa = 3.36FDD441 pKa = 6.53LNDD444 pKa = 2.91VVTYY448 pKa = 10.65YY449 pKa = 9.03RR450 pKa = 11.84TKK452 pKa = 10.78VISKK456 pKa = 9.88DD457 pKa = 3.63GNVLRR462 pKa = 11.84LDD464 pKa = 3.37IEE466 pKa = 4.57GTITAYY472 pKa = 10.08GASYY476 pKa = 11.43SNGLAWKK483 pKa = 9.92LDD485 pKa = 3.6GFAEE489 pKa = 4.47SDD491 pKa = 3.76VNLQLSRR498 pKa = 11.84VTKK501 pKa = 10.62NGQLRR506 pKa = 11.84NGISPFTGEE515 pKa = 4.62DD516 pKa = 3.16KK517 pKa = 11.11SVASPGGDD525 pKa = 2.84LVVVASVNLLDD536 pKa = 5.98DD537 pKa = 3.83IPIHH541 pKa = 5.96GEE543 pKa = 4.1CIYY546 pKa = 10.98HH547 pKa = 5.93RR548 pKa = 11.84TNPSCNPSLEE558 pKa = 4.19SSQMTFSVSLPFNDD572 pKa = 3.95GSEE575 pKa = 4.18PTVTSLLPLDD585 pKa = 3.85GHH587 pKa = 7.01DD588 pKa = 3.87PFIFASGEE596 pKa = 3.95GSYY599 pKa = 10.25HH600 pKa = 6.99GEE602 pKa = 3.98IFTVPPGKK610 pKa = 10.14DD611 pKa = 3.62LEE613 pKa = 4.21IHH615 pKa = 5.98TADD618 pKa = 4.54FPPTSRR624 pKa = 11.84GTLVSTFYY632 pKa = 10.79EE633 pKa = 4.28QADD636 pKa = 3.87DD637 pKa = 4.81RR638 pKa = 11.84SDD640 pKa = 3.59PSISKK645 pKa = 10.4YY646 pKa = 10.65YY647 pKa = 9.36RR648 pKa = 11.84TDD650 pKa = 3.2GNLPWGIIIPSIWNHH665 pKa = 5.04PSEE668 pKa = 4.83YY669 pKa = 9.31IDD671 pKa = 3.95VSVAYY676 pKa = 9.5PDD678 pKa = 3.6FAEE681 pKa = 4.3WAVSGGSSKK690 pKa = 9.13PTWYY694 pKa = 10.66LNPEE698 pKa = 4.28SSQTWTTSDD707 pKa = 2.96
Molecular weight: 76.2 kDa
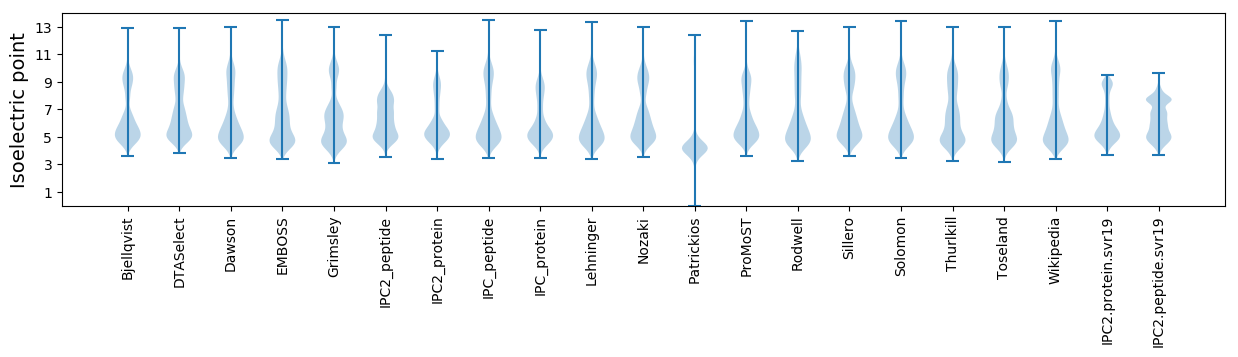
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U3C526|U3C526_9VIBR Inosine/xanthosine triphosphatase OS=Vibrio azureus NBRC 104587 OX=1219077 GN=VAZ01S_045_00470 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.5RR3 pKa = 11.84TFQPTVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44NGRR28 pKa = 11.84KK29 pKa = 9.39VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LSKK44 pKa = 10.83
MM1 pKa = 7.45KK2 pKa = 9.5RR3 pKa = 11.84TFQPTVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44NGRR28 pKa = 11.84KK29 pKa = 9.39VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LSKK44 pKa = 10.83
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1329685 |
37 |
3512 |
320.6 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.997 ± 0.04 | 1.074 ± 0.013 |
5.511 ± 0.03 | 6.453 ± 0.038 |
4.226 ± 0.027 | 6.517 ± 0.034 |
2.31 ± 0.021 | 6.443 ± 0.032 |
5.632 ± 0.036 | 10.185 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.609 ± 0.02 | 4.543 ± 0.03 |
3.866 ± 0.023 | 4.751 ± 0.036 |
4.452 ± 0.029 | 6.932 ± 0.035 |
5.386 ± 0.026 | 6.695 ± 0.029 |
1.238 ± 0.014 | 3.179 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
