
Oceanicola sp. 22II-s10i
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Oceanicola; unclassified Oceanicola
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
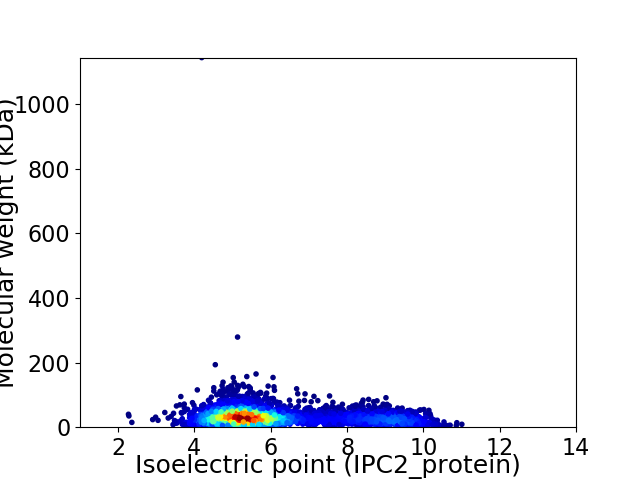
Virtual 2D-PAGE plot for 4471 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A254QXC6|A0A254QXC6_9RHOB Luciferase OS=Oceanicola sp. 22II-s10i OX=1317116 GN=ATO6_22275 PE=4 SV=1
MM1 pKa = 7.78IYY3 pKa = 10.76GEE5 pKa = 4.04QSYY8 pKa = 7.85ATSTSHH14 pKa = 7.71DD15 pKa = 3.92MIQLYY20 pKa = 10.39MNFFNMADD28 pKa = 3.28ARR30 pKa = 11.84GAKK33 pKa = 9.67AVEE36 pKa = 5.17DD37 pKa = 3.94INVMGNVLNGASVNGNALDD56 pKa = 3.57NEE58 pKa = 4.25MRR60 pKa = 11.84GNVGNDD66 pKa = 3.32TLSGGSGNDD75 pKa = 3.3EE76 pKa = 3.72IFGKK80 pKa = 10.69GGNDD84 pKa = 3.3KK85 pKa = 11.17LSGGIGNDD93 pKa = 3.63FLSGDD98 pKa = 4.02AGNDD102 pKa = 3.9FIDD105 pKa = 4.62GGNDD109 pKa = 2.71QDD111 pKa = 4.31TIYY114 pKa = 10.8GGSGNDD120 pKa = 3.85SLWGASGNDD129 pKa = 3.46VIYY132 pKa = 10.56GGSEE136 pKa = 4.01DD137 pKa = 5.31DD138 pKa = 3.76VLKK141 pKa = 11.42GEE143 pKa = 5.32DD144 pKa = 3.76GNDD147 pKa = 3.54YY148 pKa = 11.13LDD150 pKa = 4.27GGSGFDD156 pKa = 4.78RR157 pKa = 11.84MDD159 pKa = 3.54GGAGNDD165 pKa = 3.72TLKK168 pKa = 10.91GGEE171 pKa = 4.27DD172 pKa = 3.34MAGGEE177 pKa = 4.59GNDD180 pKa = 3.32TFYY183 pKa = 11.55VVDD186 pKa = 4.25DD187 pKa = 4.14AQIVTEE193 pKa = 4.63ALNEE197 pKa = 4.2GQDD200 pKa = 3.74VVRR203 pKa = 11.84TTLTEE208 pKa = 3.96YY209 pKa = 10.65TLGANLEE216 pKa = 4.15EE217 pKa = 4.53LVYY220 pKa = 10.79EE221 pKa = 4.57GDD223 pKa = 3.34VSATVSFEE231 pKa = 4.15GNASDD236 pKa = 4.67NYY238 pKa = 10.04ISARR242 pKa = 11.84NSAHH246 pKa = 6.64AILWGGDD253 pKa = 3.0GWDD256 pKa = 4.74DD257 pKa = 3.66LHH259 pKa = 7.54GSNTGYY265 pKa = 10.71DD266 pKa = 3.46QLYY269 pKa = 9.86GGSGDD274 pKa = 3.86DD275 pKa = 3.68VIYY278 pKa = 10.21GYY280 pKa = 11.23GGDD283 pKa = 4.18DD284 pKa = 4.97FINGGTGHH292 pKa = 7.72DD293 pKa = 4.02YY294 pKa = 10.73MRR296 pKa = 11.84GGTGNDD302 pKa = 3.26TYY304 pKa = 11.69VVDD307 pKa = 4.42SNLDD311 pKa = 3.79RR312 pKa = 11.84IIEE315 pKa = 4.18YY316 pKa = 10.89ANEE319 pKa = 4.7GYY321 pKa = 10.65DD322 pKa = 3.0IARR325 pKa = 11.84ITNQYY330 pKa = 10.35FKK332 pKa = 11.21LLDD335 pKa = 3.36TSMVEE340 pKa = 3.99EE341 pKa = 4.58LQATTNNGHH350 pKa = 6.5MIAGNMYY357 pKa = 9.86ANTIRR362 pKa = 11.84GLNGEE367 pKa = 4.68DD368 pKa = 3.35VLQGFDD374 pKa = 3.54GNDD377 pKa = 3.25YY378 pKa = 11.25LVGNGGNDD386 pKa = 3.46SLDD389 pKa = 3.76GGSGNDD395 pKa = 3.58TMLGGYY401 pKa = 10.56GNDD404 pKa = 3.86TYY406 pKa = 10.71WIDD409 pKa = 3.44SQYY412 pKa = 11.76DD413 pKa = 3.52KK414 pKa = 10.89IVEE417 pKa = 4.08YY418 pKa = 10.97SSGGFDD424 pKa = 2.85TAYY427 pKa = 9.81IKK429 pKa = 10.77NSYY432 pKa = 10.21YY433 pKa = 11.19SFIGSEE439 pKa = 3.89NVEE442 pKa = 3.95RR443 pKa = 11.84MIASSNWNTTIHH455 pKa = 5.88GTNGHH460 pKa = 5.26NRR462 pKa = 11.84ILGGAKK468 pKa = 9.71NDD470 pKa = 3.9LLSGRR475 pKa = 11.84GGNDD479 pKa = 3.26NIHH482 pKa = 6.62GGGGADD488 pKa = 2.71RR489 pKa = 11.84VYY491 pKa = 11.13GGTGNDD497 pKa = 3.15QFIFQNNRR505 pKa = 11.84SNTGSKK511 pKa = 10.88VMDD514 pKa = 4.35FVHH517 pKa = 7.19GEE519 pKa = 4.04DD520 pKa = 5.34KK521 pKa = 10.81IVLDD525 pKa = 6.01SDD527 pKa = 4.76GQWGWAAMGNGTVVIQSGNGGGSITVYY554 pKa = 11.14ADD556 pKa = 3.0NLYY559 pKa = 10.53YY560 pKa = 10.87GSDD563 pKa = 3.35IQLMSDD569 pKa = 3.27STIANLII576 pKa = 3.93
MM1 pKa = 7.78IYY3 pKa = 10.76GEE5 pKa = 4.04QSYY8 pKa = 7.85ATSTSHH14 pKa = 7.71DD15 pKa = 3.92MIQLYY20 pKa = 10.39MNFFNMADD28 pKa = 3.28ARR30 pKa = 11.84GAKK33 pKa = 9.67AVEE36 pKa = 5.17DD37 pKa = 3.94INVMGNVLNGASVNGNALDD56 pKa = 3.57NEE58 pKa = 4.25MRR60 pKa = 11.84GNVGNDD66 pKa = 3.32TLSGGSGNDD75 pKa = 3.3EE76 pKa = 3.72IFGKK80 pKa = 10.69GGNDD84 pKa = 3.3KK85 pKa = 11.17LSGGIGNDD93 pKa = 3.63FLSGDD98 pKa = 4.02AGNDD102 pKa = 3.9FIDD105 pKa = 4.62GGNDD109 pKa = 2.71QDD111 pKa = 4.31TIYY114 pKa = 10.8GGSGNDD120 pKa = 3.85SLWGASGNDD129 pKa = 3.46VIYY132 pKa = 10.56GGSEE136 pKa = 4.01DD137 pKa = 5.31DD138 pKa = 3.76VLKK141 pKa = 11.42GEE143 pKa = 5.32DD144 pKa = 3.76GNDD147 pKa = 3.54YY148 pKa = 11.13LDD150 pKa = 4.27GGSGFDD156 pKa = 4.78RR157 pKa = 11.84MDD159 pKa = 3.54GGAGNDD165 pKa = 3.72TLKK168 pKa = 10.91GGEE171 pKa = 4.27DD172 pKa = 3.34MAGGEE177 pKa = 4.59GNDD180 pKa = 3.32TFYY183 pKa = 11.55VVDD186 pKa = 4.25DD187 pKa = 4.14AQIVTEE193 pKa = 4.63ALNEE197 pKa = 4.2GQDD200 pKa = 3.74VVRR203 pKa = 11.84TTLTEE208 pKa = 3.96YY209 pKa = 10.65TLGANLEE216 pKa = 4.15EE217 pKa = 4.53LVYY220 pKa = 10.79EE221 pKa = 4.57GDD223 pKa = 3.34VSATVSFEE231 pKa = 4.15GNASDD236 pKa = 4.67NYY238 pKa = 10.04ISARR242 pKa = 11.84NSAHH246 pKa = 6.64AILWGGDD253 pKa = 3.0GWDD256 pKa = 4.74DD257 pKa = 3.66LHH259 pKa = 7.54GSNTGYY265 pKa = 10.71DD266 pKa = 3.46QLYY269 pKa = 9.86GGSGDD274 pKa = 3.86DD275 pKa = 3.68VIYY278 pKa = 10.21GYY280 pKa = 11.23GGDD283 pKa = 4.18DD284 pKa = 4.97FINGGTGHH292 pKa = 7.72DD293 pKa = 4.02YY294 pKa = 10.73MRR296 pKa = 11.84GGTGNDD302 pKa = 3.26TYY304 pKa = 11.69VVDD307 pKa = 4.42SNLDD311 pKa = 3.79RR312 pKa = 11.84IIEE315 pKa = 4.18YY316 pKa = 10.89ANEE319 pKa = 4.7GYY321 pKa = 10.65DD322 pKa = 3.0IARR325 pKa = 11.84ITNQYY330 pKa = 10.35FKK332 pKa = 11.21LLDD335 pKa = 3.36TSMVEE340 pKa = 3.99EE341 pKa = 4.58LQATTNNGHH350 pKa = 6.5MIAGNMYY357 pKa = 9.86ANTIRR362 pKa = 11.84GLNGEE367 pKa = 4.68DD368 pKa = 3.35VLQGFDD374 pKa = 3.54GNDD377 pKa = 3.25YY378 pKa = 11.25LVGNGGNDD386 pKa = 3.46SLDD389 pKa = 3.76GGSGNDD395 pKa = 3.58TMLGGYY401 pKa = 10.56GNDD404 pKa = 3.86TYY406 pKa = 10.71WIDD409 pKa = 3.44SQYY412 pKa = 11.76DD413 pKa = 3.52KK414 pKa = 10.89IVEE417 pKa = 4.08YY418 pKa = 10.97SSGGFDD424 pKa = 2.85TAYY427 pKa = 9.81IKK429 pKa = 10.77NSYY432 pKa = 10.21YY433 pKa = 11.19SFIGSEE439 pKa = 3.89NVEE442 pKa = 3.95RR443 pKa = 11.84MIASSNWNTTIHH455 pKa = 5.88GTNGHH460 pKa = 5.26NRR462 pKa = 11.84ILGGAKK468 pKa = 9.71NDD470 pKa = 3.9LLSGRR475 pKa = 11.84GGNDD479 pKa = 3.26NIHH482 pKa = 6.62GGGGADD488 pKa = 2.71RR489 pKa = 11.84VYY491 pKa = 11.13GGTGNDD497 pKa = 3.15QFIFQNNRR505 pKa = 11.84SNTGSKK511 pKa = 10.88VMDD514 pKa = 4.35FVHH517 pKa = 7.19GEE519 pKa = 4.04DD520 pKa = 5.34KK521 pKa = 10.81IVLDD525 pKa = 6.01SDD527 pKa = 4.76GQWGWAAMGNGTVVIQSGNGGGSITVYY554 pKa = 11.14ADD556 pKa = 3.0NLYY559 pKa = 10.53YY560 pKa = 10.87GSDD563 pKa = 3.35IQLMSDD569 pKa = 3.27STIANLII576 pKa = 3.93
Molecular weight: 60.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A254R370|A0A254R370_9RHOB Uncharacterized protein OS=Oceanicola sp. 22II-s10i OX=1317116 GN=ATO6_17525 PE=4 SV=1
MM1 pKa = 7.48PAAHH5 pKa = 6.95SFRR8 pKa = 11.84TRR10 pKa = 11.84LRR12 pKa = 11.84RR13 pKa = 11.84FQVQARR19 pKa = 11.84RR20 pKa = 11.84SIPPGLRR27 pKa = 11.84LVLGILLIIGGFLGFLPILGFWMLPLGIAIAALDD61 pKa = 3.96VVPVWRR67 pKa = 11.84RR68 pKa = 11.84MVSLRR73 pKa = 11.84RR74 pKa = 11.84PRR76 pKa = 11.84RR77 pKa = 11.84PRR79 pKa = 3.53
MM1 pKa = 7.48PAAHH5 pKa = 6.95SFRR8 pKa = 11.84TRR10 pKa = 11.84LRR12 pKa = 11.84RR13 pKa = 11.84FQVQARR19 pKa = 11.84RR20 pKa = 11.84SIPPGLRR27 pKa = 11.84LVLGILLIIGGFLGFLPILGFWMLPLGIAIAALDD61 pKa = 3.96VVPVWRR67 pKa = 11.84RR68 pKa = 11.84MVSLRR73 pKa = 11.84RR74 pKa = 11.84PRR76 pKa = 11.84RR77 pKa = 11.84PRR79 pKa = 3.53
Molecular weight: 9.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1400842 |
32 |
10931 |
313.3 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.544 ± 0.046 | 0.865 ± 0.013 |
6.134 ± 0.03 | 5.964 ± 0.03 |
3.639 ± 0.023 | 9.079 ± 0.036 |
1.994 ± 0.018 | 5.112 ± 0.028 |
2.961 ± 0.036 | 9.814 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.895 ± 0.026 | 2.426 ± 0.022 |
5.199 ± 0.024 | 2.885 ± 0.017 |
7.025 ± 0.038 | 4.914 ± 0.024 |
5.535 ± 0.039 | 7.412 ± 0.029 |
1.388 ± 0.015 | 2.216 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
