
Verrucomicrobiaceae bacterium
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Verrucomicrobiae; Verrucomicrobiales; Verrucomicrobiaceae; unclassified Verrucomicrobiaceae
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
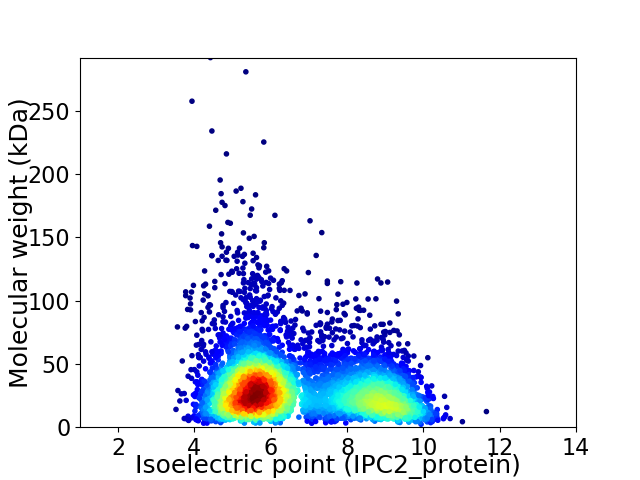
Virtual 2D-PAGE plot for 5527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
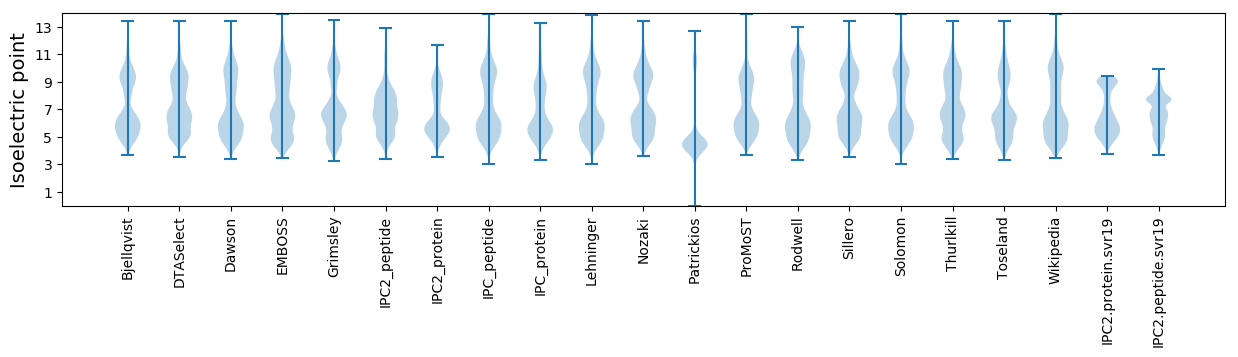
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q2XX62|A0A4Q2XX62_9BACT D-arabino 3-hexulose 6-phosphate aldehyde lyase OS=Verrucomicrobiaceae bacterium OX=2026800 GN=EOP86_22540 PE=4 SV=1
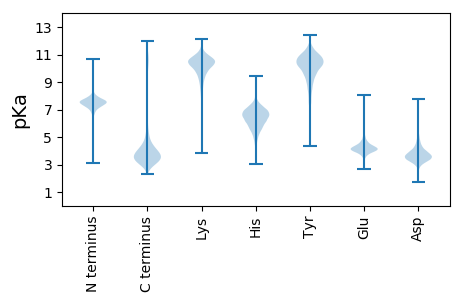
AA1 pKa = 7.4TGVNGDD7 pKa = 3.43QADD10 pKa = 3.95NSAHH14 pKa = 5.7QSGAAYY20 pKa = 9.57IFTVADD26 pKa = 3.56YY27 pKa = 9.96EE28 pKa = 4.43YY29 pKa = 11.25SLLTTAVNGTVTGAGEE45 pKa = 4.17YY46 pKa = 10.94AFGTTATLTATPDD59 pKa = 2.95AGYY62 pKa = 11.12VFTGWTGDD70 pKa = 3.73AGGTDD75 pKa = 3.38NPLPVLMDD83 pKa = 3.67SDD85 pKa = 3.87RR86 pKa = 11.84TIGAVFGPDD95 pKa = 3.3SSDD98 pKa = 3.38ADD100 pKa = 3.82GDD102 pKa = 4.03GLSAYY107 pKa = 9.89LEE109 pKa = 3.89RR110 pKa = 11.84VVYY113 pKa = 8.04GTNPLLADD121 pKa = 3.57TDD123 pKa = 4.25GDD125 pKa = 4.13SLSDD129 pKa = 3.3KK130 pKa = 11.31GEE132 pKa = 4.07VEE134 pKa = 3.85ITLTNPLVNDD144 pKa = 3.78SDD146 pKa = 4.54GDD148 pKa = 3.85WVLDD152 pKa = 3.94ADD154 pKa = 4.84EE155 pKa = 6.14DD156 pKa = 4.49PDD158 pKa = 5.55GDD160 pKa = 3.98GFTNGQEE167 pKa = 3.96LTLFLTDD174 pKa = 4.31PNSGADD180 pKa = 3.6CFTTALEE187 pKa = 4.41YY188 pKa = 9.62TASAHH193 pKa = 6.14SLTFATVAGRR203 pKa = 11.84TYY205 pKa = 10.28QVEE208 pKa = 4.25RR209 pKa = 11.84ALKK212 pKa = 10.2PGDD215 pKa = 3.3PAAWSEE221 pKa = 4.13RR222 pKa = 11.84FSFIASGGAVTVPLGPPSSTRR243 pKa = 11.84WFYY246 pKa = 10.99RR247 pKa = 11.84VRR249 pKa = 11.84VSLNN253 pKa = 2.94
AA1 pKa = 7.4TGVNGDD7 pKa = 3.43QADD10 pKa = 3.95NSAHH14 pKa = 5.7QSGAAYY20 pKa = 9.57IFTVADD26 pKa = 3.56YY27 pKa = 9.96EE28 pKa = 4.43YY29 pKa = 11.25SLLTTAVNGTVTGAGEE45 pKa = 4.17YY46 pKa = 10.94AFGTTATLTATPDD59 pKa = 2.95AGYY62 pKa = 11.12VFTGWTGDD70 pKa = 3.73AGGTDD75 pKa = 3.38NPLPVLMDD83 pKa = 3.67SDD85 pKa = 3.87RR86 pKa = 11.84TIGAVFGPDD95 pKa = 3.3SSDD98 pKa = 3.38ADD100 pKa = 3.82GDD102 pKa = 4.03GLSAYY107 pKa = 9.89LEE109 pKa = 3.89RR110 pKa = 11.84VVYY113 pKa = 8.04GTNPLLADD121 pKa = 3.57TDD123 pKa = 4.25GDD125 pKa = 4.13SLSDD129 pKa = 3.3KK130 pKa = 11.31GEE132 pKa = 4.07VEE134 pKa = 3.85ITLTNPLVNDD144 pKa = 3.78SDD146 pKa = 4.54GDD148 pKa = 3.85WVLDD152 pKa = 3.94ADD154 pKa = 4.84EE155 pKa = 6.14DD156 pKa = 4.49PDD158 pKa = 5.55GDD160 pKa = 3.98GFTNGQEE167 pKa = 3.96LTLFLTDD174 pKa = 4.31PNSGADD180 pKa = 3.6CFTTALEE187 pKa = 4.41YY188 pKa = 9.62TASAHH193 pKa = 6.14SLTFATVAGRR203 pKa = 11.84TYY205 pKa = 10.28QVEE208 pKa = 4.25RR209 pKa = 11.84ALKK212 pKa = 10.2PGDD215 pKa = 3.3PAAWSEE221 pKa = 4.13RR222 pKa = 11.84FSFIASGGAVTVPLGPPSSTRR243 pKa = 11.84WFYY246 pKa = 10.99RR247 pKa = 11.84VRR249 pKa = 11.84VSLNN253 pKa = 2.94
Molecular weight: 26.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q2XNC4|A0A4Q2XNC4_9BACT DUF1778 domain-containing protein OS=Verrucomicrobiaceae bacterium OX=2026800 GN=EOP86_26125 PE=4 SV=1
MM1 pKa = 7.94PSRR4 pKa = 11.84AAARR8 pKa = 11.84RR9 pKa = 11.84VVVRR13 pKa = 11.84VPVKK17 pKa = 10.0AVRR20 pKa = 11.84NVRR23 pKa = 11.84RR24 pKa = 11.84APKK27 pKa = 8.34VAHH30 pKa = 4.97VAGRR34 pKa = 11.84VVNAAAPQPRR44 pKa = 11.84VSRR47 pKa = 11.84VRR49 pKa = 11.84LRR51 pKa = 11.84RR52 pKa = 11.84LLVPPLLLPNRR63 pKa = 11.84LRR65 pKa = 11.84RR66 pKa = 11.84VQPLRR71 pKa = 11.84QRR73 pKa = 11.84LPPRR77 pKa = 11.84VNPLHH82 pKa = 7.18RR83 pKa = 11.84LRR85 pKa = 11.84HH86 pKa = 5.47LPRR89 pKa = 11.84ALPRR93 pKa = 11.84HH94 pKa = 5.81RR95 pKa = 11.84RR96 pKa = 11.84PLLLRR101 pKa = 11.84RR102 pKa = 11.84LRR104 pKa = 11.84LPP106 pKa = 3.56
MM1 pKa = 7.94PSRR4 pKa = 11.84AAARR8 pKa = 11.84RR9 pKa = 11.84VVVRR13 pKa = 11.84VPVKK17 pKa = 10.0AVRR20 pKa = 11.84NVRR23 pKa = 11.84RR24 pKa = 11.84APKK27 pKa = 8.34VAHH30 pKa = 4.97VAGRR34 pKa = 11.84VVNAAAPQPRR44 pKa = 11.84VSRR47 pKa = 11.84VRR49 pKa = 11.84LRR51 pKa = 11.84RR52 pKa = 11.84LLVPPLLLPNRR63 pKa = 11.84LRR65 pKa = 11.84RR66 pKa = 11.84VQPLRR71 pKa = 11.84QRR73 pKa = 11.84LPPRR77 pKa = 11.84VNPLHH82 pKa = 7.18RR83 pKa = 11.84LRR85 pKa = 11.84HH86 pKa = 5.47LPRR89 pKa = 11.84ALPRR93 pKa = 11.84HH94 pKa = 5.81RR95 pKa = 11.84RR96 pKa = 11.84PLLLRR101 pKa = 11.84RR102 pKa = 11.84LRR104 pKa = 11.84LPP106 pKa = 3.56
Molecular weight: 12.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1744395 |
21 |
2852 |
315.6 |
34.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.309 ± 0.046 | 0.96 ± 0.013 |
5.294 ± 0.027 | 5.789 ± 0.032 |
3.757 ± 0.018 | 8.812 ± 0.038 |
2.137 ± 0.018 | 4.275 ± 0.028 |
3.57 ± 0.032 | 10.28 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.188 ± 0.016 | 2.911 ± 0.025 |
6.169 ± 0.031 | 3.24 ± 0.017 |
6.825 ± 0.035 | 6.098 ± 0.028 |
5.811 ± 0.033 | 6.712 ± 0.029 |
1.7 ± 0.017 | 2.165 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
