
Pararhodospirillum oryzae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Pararhodospirillum
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
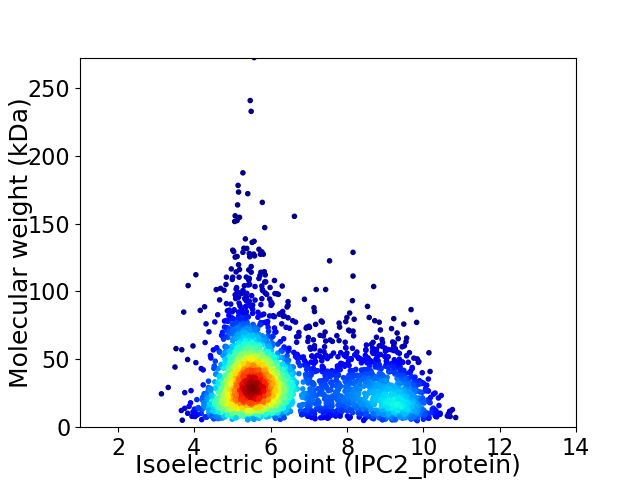
Virtual 2D-PAGE plot for 3199 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A512H4N5|A0A512H4N5_9PROT Uncharacterized protein OS=Pararhodospirillum oryzae OX=478448 GN=ROR02_04710 PE=3 SV=1
MM1 pKa = 6.61TTITSSSYY9 pKa = 7.72VTGSSGTTYY18 pKa = 11.16LSGLSDD24 pKa = 4.87LDD26 pKa = 3.55TSALIEE32 pKa = 4.19AAVEE36 pKa = 4.26AKK38 pKa = 8.25MQPAYY43 pKa = 10.74RR44 pKa = 11.84LDD46 pKa = 3.92DD47 pKa = 4.8KK48 pKa = 11.32ISALEE53 pKa = 4.15TEE55 pKa = 4.62EE56 pKa = 4.6SAFSEE61 pKa = 4.43MTSLLSAVSEE71 pKa = 4.05ALEE74 pKa = 4.19PLASASVLSSSSDD87 pKa = 3.33TTSAFAGRR95 pKa = 11.84SAYY98 pKa = 8.46LTSSLDD104 pKa = 3.33TPANYY109 pKa = 9.97VGLTVTDD116 pKa = 3.84EE117 pKa = 4.44ADD119 pKa = 3.24LGNYY123 pKa = 7.14TLSVEE128 pKa = 4.71QLATTQKK135 pKa = 10.29VASQGLSAGTALGVDD150 pKa = 4.33GSFTLQADD158 pKa = 3.61DD159 pKa = 4.0GTAVDD164 pKa = 3.72ITVDD168 pKa = 3.36ASMTASDD175 pKa = 3.09IAEE178 pKa = 4.96AINEE182 pKa = 3.91QSATSGVVATVVRR195 pKa = 11.84TGSNEE200 pKa = 3.62QTLVLSSVNTGQAFTLTEE218 pKa = 3.97TSGTAGQALGLVDD231 pKa = 4.23SSGDD235 pKa = 3.43FVKK238 pKa = 10.43PVQEE242 pKa = 4.03AQQAILTLDD251 pKa = 3.67GVTVTSDD258 pKa = 3.26SNDD261 pKa = 3.2IKK263 pKa = 11.26DD264 pKa = 4.19LLPGVSVNLYY274 pKa = 9.88AATGGEE280 pKa = 4.38TIAVEE285 pKa = 4.22VGQDD289 pKa = 3.23LGAALDD295 pKa = 4.54AIEE298 pKa = 5.08AFVEE302 pKa = 4.51AYY304 pKa = 9.44NAYY307 pKa = 10.32RR308 pKa = 11.84EE309 pKa = 4.21FALTQQATDD318 pKa = 3.39EE319 pKa = 4.55SGAADD324 pKa = 3.76DD325 pKa = 5.34AVLFGDD331 pKa = 3.98STLKK335 pKa = 10.69GVNAALYY342 pKa = 9.68SALNTSVTVDD352 pKa = 3.0GTTYY356 pKa = 10.09TLSDD360 pKa = 3.41MGLSFNDD367 pKa = 4.18DD368 pKa = 3.05NTLALDD374 pKa = 3.81GDD376 pKa = 4.65VIEE379 pKa = 5.91EE380 pKa = 4.14MLLQNPDD387 pKa = 2.92VVEE390 pKa = 4.84AFFCSSVSCDD400 pKa = 3.26SSSLGVVSLPATMPSGTYY418 pKa = 9.62TLDD421 pKa = 3.19IDD423 pKa = 3.82VDD425 pKa = 3.81EE426 pKa = 4.81ATGKK430 pKa = 9.91IASATLNGVALTVSGTTLSGPDD452 pKa = 3.3GSIYY456 pKa = 10.26EE457 pKa = 4.6DD458 pKa = 2.99LWMGYY463 pKa = 9.97SGTGDD468 pKa = 3.3ATITLAVSTGLADD481 pKa = 4.42ALTTAIDD488 pKa = 4.45DD489 pKa = 3.84YY490 pKa = 11.58TNAGDD495 pKa = 4.05GRR497 pKa = 11.84ITTRR501 pKa = 11.84LSSIADD507 pKa = 4.56DD508 pKa = 3.37ITGKK512 pKa = 8.03QEE514 pKa = 3.54KK515 pKa = 9.29RR516 pKa = 11.84DD517 pKa = 4.15RR518 pKa = 11.84IAEE521 pKa = 4.0RR522 pKa = 11.84AGDD525 pKa = 3.69YY526 pKa = 9.63EE527 pKa = 4.27EE528 pKa = 6.29RR529 pKa = 11.84MIAYY533 pKa = 7.77YY534 pKa = 10.82AKK536 pKa = 10.64LEE538 pKa = 4.21EE539 pKa = 4.57QISMADD545 pKa = 2.92IRR547 pKa = 11.84LQQIEE552 pKa = 4.24ALFYY556 pKa = 11.29GDD558 pKa = 5.55DD559 pKa = 3.79DD560 pKa = 4.03
MM1 pKa = 6.61TTITSSSYY9 pKa = 7.72VTGSSGTTYY18 pKa = 11.16LSGLSDD24 pKa = 4.87LDD26 pKa = 3.55TSALIEE32 pKa = 4.19AAVEE36 pKa = 4.26AKK38 pKa = 8.25MQPAYY43 pKa = 10.74RR44 pKa = 11.84LDD46 pKa = 3.92DD47 pKa = 4.8KK48 pKa = 11.32ISALEE53 pKa = 4.15TEE55 pKa = 4.62EE56 pKa = 4.6SAFSEE61 pKa = 4.43MTSLLSAVSEE71 pKa = 4.05ALEE74 pKa = 4.19PLASASVLSSSSDD87 pKa = 3.33TTSAFAGRR95 pKa = 11.84SAYY98 pKa = 8.46LTSSLDD104 pKa = 3.33TPANYY109 pKa = 9.97VGLTVTDD116 pKa = 3.84EE117 pKa = 4.44ADD119 pKa = 3.24LGNYY123 pKa = 7.14TLSVEE128 pKa = 4.71QLATTQKK135 pKa = 10.29VASQGLSAGTALGVDD150 pKa = 4.33GSFTLQADD158 pKa = 3.61DD159 pKa = 4.0GTAVDD164 pKa = 3.72ITVDD168 pKa = 3.36ASMTASDD175 pKa = 3.09IAEE178 pKa = 4.96AINEE182 pKa = 3.91QSATSGVVATVVRR195 pKa = 11.84TGSNEE200 pKa = 3.62QTLVLSSVNTGQAFTLTEE218 pKa = 3.97TSGTAGQALGLVDD231 pKa = 4.23SSGDD235 pKa = 3.43FVKK238 pKa = 10.43PVQEE242 pKa = 4.03AQQAILTLDD251 pKa = 3.67GVTVTSDD258 pKa = 3.26SNDD261 pKa = 3.2IKK263 pKa = 11.26DD264 pKa = 4.19LLPGVSVNLYY274 pKa = 9.88AATGGEE280 pKa = 4.38TIAVEE285 pKa = 4.22VGQDD289 pKa = 3.23LGAALDD295 pKa = 4.54AIEE298 pKa = 5.08AFVEE302 pKa = 4.51AYY304 pKa = 9.44NAYY307 pKa = 10.32RR308 pKa = 11.84EE309 pKa = 4.21FALTQQATDD318 pKa = 3.39EE319 pKa = 4.55SGAADD324 pKa = 3.76DD325 pKa = 5.34AVLFGDD331 pKa = 3.98STLKK335 pKa = 10.69GVNAALYY342 pKa = 9.68SALNTSVTVDD352 pKa = 3.0GTTYY356 pKa = 10.09TLSDD360 pKa = 3.41MGLSFNDD367 pKa = 4.18DD368 pKa = 3.05NTLALDD374 pKa = 3.81GDD376 pKa = 4.65VIEE379 pKa = 5.91EE380 pKa = 4.14MLLQNPDD387 pKa = 2.92VVEE390 pKa = 4.84AFFCSSVSCDD400 pKa = 3.26SSSLGVVSLPATMPSGTYY418 pKa = 9.62TLDD421 pKa = 3.19IDD423 pKa = 3.82VDD425 pKa = 3.81EE426 pKa = 4.81ATGKK430 pKa = 9.91IASATLNGVALTVSGTTLSGPDD452 pKa = 3.3GSIYY456 pKa = 10.26EE457 pKa = 4.6DD458 pKa = 2.99LWMGYY463 pKa = 9.97SGTGDD468 pKa = 3.3ATITLAVSTGLADD481 pKa = 4.42ALTTAIDD488 pKa = 4.45DD489 pKa = 3.84YY490 pKa = 11.58TNAGDD495 pKa = 4.05GRR497 pKa = 11.84ITTRR501 pKa = 11.84LSSIADD507 pKa = 4.56DD508 pKa = 3.37ITGKK512 pKa = 8.03QEE514 pKa = 3.54KK515 pKa = 9.29RR516 pKa = 11.84DD517 pKa = 4.15RR518 pKa = 11.84IAEE521 pKa = 4.0RR522 pKa = 11.84AGDD525 pKa = 3.69YY526 pKa = 9.63EE527 pKa = 4.27EE528 pKa = 6.29RR529 pKa = 11.84MIAYY533 pKa = 7.77YY534 pKa = 10.82AKK536 pKa = 10.64LEE538 pKa = 4.21EE539 pKa = 4.57QISMADD545 pKa = 2.92IRR547 pKa = 11.84LQQIEE552 pKa = 4.24ALFYY556 pKa = 11.29GDD558 pKa = 5.55DD559 pKa = 3.79DD560 pKa = 4.03
Molecular weight: 57.86 kDa
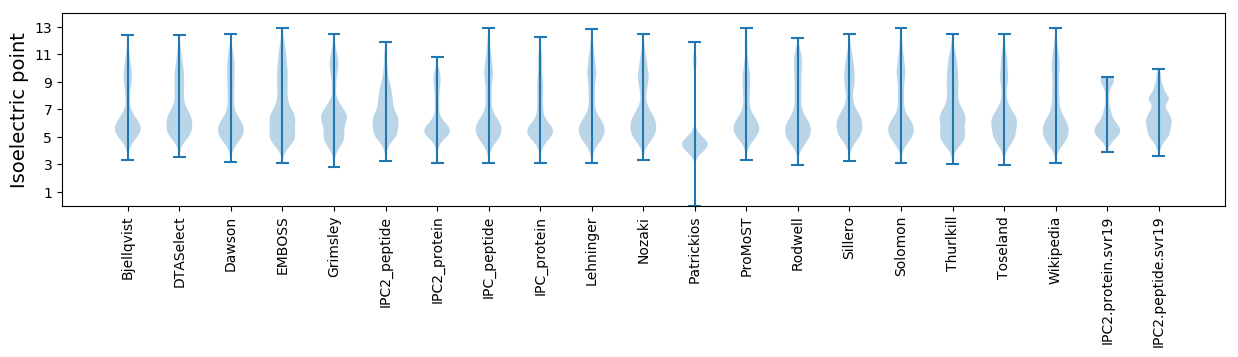
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A512H3N1|A0A512H3N1_9PROT Peptide chain release factor 1 OS=Pararhodospirillum oryzae OX=478448 GN=prfA PE=3 SV=1
MM1 pKa = 7.5SALSLISRR9 pKa = 11.84HH10 pKa = 6.12PVATWRR16 pKa = 11.84LWHH19 pKa = 6.74AGLVGSGLVAYY30 pKa = 7.49VTADD34 pKa = 3.21EE35 pKa = 4.4DD36 pKa = 4.47TYY38 pKa = 11.93ALHH41 pKa = 6.24QFSGYY46 pKa = 10.15LFAVLAVGRR55 pKa = 11.84LIVAAAPGIRR65 pKa = 11.84GPWRR69 pKa = 11.84LPQPGGKK76 pKa = 9.36AAAPARR82 pKa = 11.84LTRR85 pKa = 11.84GAMFQRR91 pKa = 11.84VRR93 pKa = 11.84PLMTAVLIGVLLAAGLSGVGADD115 pKa = 3.42WIHH118 pKa = 6.59RR119 pKa = 11.84LEE121 pKa = 4.42KK122 pKa = 10.28LHH124 pKa = 6.24EE125 pKa = 4.55AVGEE129 pKa = 3.99ATPWVVLAHH138 pKa = 5.41VAVVVALLARR148 pKa = 11.84LPQGLRR154 pKa = 11.84RR155 pKa = 11.84MATPQRR161 pKa = 11.84ALSALAAAGLLATLMGAAPPARR183 pKa = 11.84AADD186 pKa = 3.83PQQALLGVFTEE197 pKa = 4.48RR198 pKa = 11.84AKK200 pKa = 10.94AATPGFAGFDD210 pKa = 3.81PKK212 pKa = 10.63RR213 pKa = 11.84GEE215 pKa = 4.02ALFLSRR221 pKa = 11.84NTANPEE227 pKa = 4.02LPSCASCHH235 pKa = 5.34TQDD238 pKa = 3.27PRR240 pKa = 11.84ASGRR244 pKa = 11.84HH245 pKa = 5.11AKK247 pKa = 9.04TGRR250 pKa = 11.84AIDD253 pKa = 3.73PMAPSVNRR261 pKa = 11.84EE262 pKa = 3.76RR263 pKa = 11.84FTDD266 pKa = 4.18LAQVDD271 pKa = 3.74KK272 pKa = 11.14RR273 pKa = 11.84LGRR276 pKa = 11.84DD277 pKa = 3.27CEE279 pKa = 4.5TVLGRR284 pKa = 11.84ACTPLEE290 pKa = 3.97QGDD293 pKa = 4.3FVAFMLSRR301 pKa = 3.88
MM1 pKa = 7.5SALSLISRR9 pKa = 11.84HH10 pKa = 6.12PVATWRR16 pKa = 11.84LWHH19 pKa = 6.74AGLVGSGLVAYY30 pKa = 7.49VTADD34 pKa = 3.21EE35 pKa = 4.4DD36 pKa = 4.47TYY38 pKa = 11.93ALHH41 pKa = 6.24QFSGYY46 pKa = 10.15LFAVLAVGRR55 pKa = 11.84LIVAAAPGIRR65 pKa = 11.84GPWRR69 pKa = 11.84LPQPGGKK76 pKa = 9.36AAAPARR82 pKa = 11.84LTRR85 pKa = 11.84GAMFQRR91 pKa = 11.84VRR93 pKa = 11.84PLMTAVLIGVLLAAGLSGVGADD115 pKa = 3.42WIHH118 pKa = 6.59RR119 pKa = 11.84LEE121 pKa = 4.42KK122 pKa = 10.28LHH124 pKa = 6.24EE125 pKa = 4.55AVGEE129 pKa = 3.99ATPWVVLAHH138 pKa = 5.41VAVVVALLARR148 pKa = 11.84LPQGLRR154 pKa = 11.84RR155 pKa = 11.84MATPQRR161 pKa = 11.84ALSALAAAGLLATLMGAAPPARR183 pKa = 11.84AADD186 pKa = 3.83PQQALLGVFTEE197 pKa = 4.48RR198 pKa = 11.84AKK200 pKa = 10.94AATPGFAGFDD210 pKa = 3.81PKK212 pKa = 10.63RR213 pKa = 11.84GEE215 pKa = 4.02ALFLSRR221 pKa = 11.84NTANPEE227 pKa = 4.02LPSCASCHH235 pKa = 5.34TQDD238 pKa = 3.27PRR240 pKa = 11.84ASGRR244 pKa = 11.84HH245 pKa = 5.11AKK247 pKa = 9.04TGRR250 pKa = 11.84AIDD253 pKa = 3.73PMAPSVNRR261 pKa = 11.84EE262 pKa = 3.76RR263 pKa = 11.84FTDD266 pKa = 4.18LAQVDD271 pKa = 3.74KK272 pKa = 11.14RR273 pKa = 11.84LGRR276 pKa = 11.84DD277 pKa = 3.27CEE279 pKa = 4.5TVLGRR284 pKa = 11.84ACTPLEE290 pKa = 3.97QGDD293 pKa = 4.3FVAFMLSRR301 pKa = 3.88
Molecular weight: 31.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1075380 |
44 |
2547 |
336.2 |
36.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.258 ± 0.068 | 0.905 ± 0.015 |
5.729 ± 0.032 | 5.634 ± 0.037 |
3.202 ± 0.026 | 8.848 ± 0.045 |
2.134 ± 0.019 | 4.03 ± 0.036 |
2.276 ± 0.026 | 11.241 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.208 ± 0.019 | 2.083 ± 0.025 |
6.158 ± 0.046 | 2.952 ± 0.024 |
7.973 ± 0.04 | 4.853 ± 0.028 |
5.455 ± 0.031 | 7.924 ± 0.033 |
1.324 ± 0.016 | 1.813 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
