
Leptospira phage vb_LkmZ_Bejolso9-LE1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified dsDNA phages
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
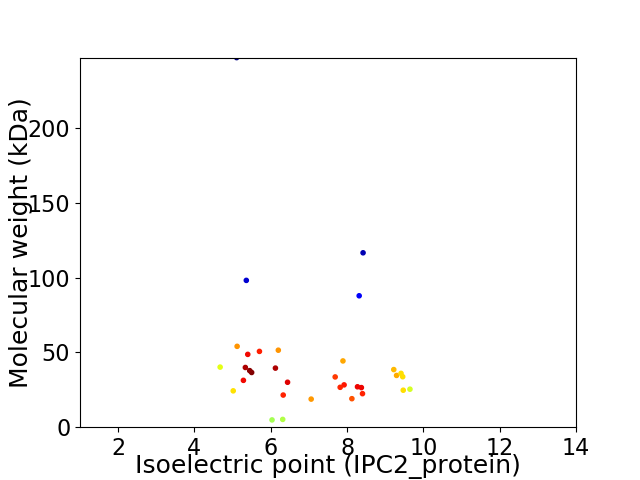
Virtual 2D-PAGE plot for 34 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5VY87|S5VY87_9VIRU Uncharacterized protein OS=Leptospira phage vb_LkmZ_Bejolso9-LE1 OX=1334244 GN=LEP1GSC052_0086 PE=4 SV=1
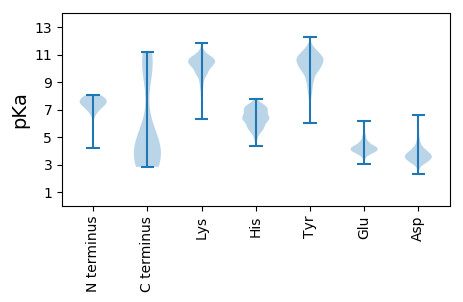
MM1 pKa = 7.39KK2 pKa = 10.57AKK4 pKa = 10.51DD5 pKa = 3.89LFFKK9 pKa = 10.52RR10 pKa = 11.84HH11 pKa = 5.63PSALKK16 pKa = 10.15NFVYY20 pKa = 9.07DD21 pKa = 3.6TLEE24 pKa = 4.1KK25 pKa = 10.75LFQNDD30 pKa = 2.4SDD32 pKa = 5.53LEE34 pKa = 4.37VEE36 pKa = 4.34TLNSIVRR43 pKa = 11.84IQKK46 pKa = 10.17EE47 pKa = 4.41SVKK50 pKa = 10.88NDD52 pKa = 4.48LIYY55 pKa = 10.98SNEE58 pKa = 3.67YY59 pKa = 10.38RR60 pKa = 11.84INQLMTSLIEE70 pKa = 3.84HH71 pKa = 6.33SKK73 pKa = 10.98DD74 pKa = 3.56FEE76 pKa = 4.37IKK78 pKa = 10.22SLSKK82 pKa = 10.4KK83 pKa = 10.06IYY85 pKa = 9.47IGVTNTYY92 pKa = 8.16TFEE95 pKa = 4.26AKK97 pKa = 10.24AVFPEE102 pKa = 4.47DD103 pKa = 3.94DD104 pKa = 3.67EE105 pKa = 4.94SLIEE109 pKa = 4.11INFGFYY115 pKa = 10.95YY116 pKa = 10.76LINLYY121 pKa = 10.78GDD123 pKa = 3.85VITRR127 pKa = 11.84LGLTIRR133 pKa = 11.84ADD135 pKa = 4.06FNEE138 pKa = 4.23SSQVLDD144 pKa = 4.85VIDD147 pKa = 4.71SDD149 pKa = 3.64IQKK152 pKa = 10.37GINAIISNFNYY163 pKa = 9.13FQKK166 pKa = 10.71KK167 pKa = 8.79IVSLEE172 pKa = 3.84KK173 pKa = 11.07GPDD176 pKa = 3.24FEE178 pKa = 5.6YY179 pKa = 10.16WGNDD183 pKa = 2.63NWNEE187 pKa = 3.81RR188 pKa = 11.84MEE190 pKa = 4.43NMAFRR195 pKa = 11.84IKK197 pKa = 10.5SIMYY201 pKa = 9.33LFVLLHH207 pKa = 5.9EE208 pKa = 5.06VGHH211 pKa = 6.86HH212 pKa = 6.65IYY214 pKa = 10.98DD215 pKa = 3.65HH216 pKa = 6.76LKK218 pKa = 10.15GFQSYY223 pKa = 10.67SLPDD227 pKa = 3.47IPNQILASIDD237 pKa = 3.45QNKK240 pKa = 9.78YY241 pKa = 10.95NIDD244 pKa = 3.55QIQEE248 pKa = 3.92FEE250 pKa = 4.41ADD252 pKa = 4.04LFAANYY258 pKa = 9.36LLNIRR263 pKa = 11.84DD264 pKa = 3.85YY265 pKa = 11.3TDD267 pKa = 4.37DD268 pKa = 4.97DD269 pKa = 5.23SILKK273 pKa = 10.53LLDD276 pKa = 3.33NTTCSLGILITLHH289 pKa = 5.69LTAGVLGDD297 pKa = 3.71IEE299 pKa = 4.66IGNEE303 pKa = 3.75NHH305 pKa = 7.0PPLSDD310 pKa = 4.49RR311 pKa = 11.84IQIIILFIQQKK322 pKa = 9.65VGQSEE327 pKa = 4.18FNEE330 pKa = 4.8IIDD333 pKa = 3.76TFQNLVYY340 pKa = 10.55RR341 pKa = 11.84IVNFSS346 pKa = 3.0
MM1 pKa = 7.39KK2 pKa = 10.57AKK4 pKa = 10.51DD5 pKa = 3.89LFFKK9 pKa = 10.52RR10 pKa = 11.84HH11 pKa = 5.63PSALKK16 pKa = 10.15NFVYY20 pKa = 9.07DD21 pKa = 3.6TLEE24 pKa = 4.1KK25 pKa = 10.75LFQNDD30 pKa = 2.4SDD32 pKa = 5.53LEE34 pKa = 4.37VEE36 pKa = 4.34TLNSIVRR43 pKa = 11.84IQKK46 pKa = 10.17EE47 pKa = 4.41SVKK50 pKa = 10.88NDD52 pKa = 4.48LIYY55 pKa = 10.98SNEE58 pKa = 3.67YY59 pKa = 10.38RR60 pKa = 11.84INQLMTSLIEE70 pKa = 3.84HH71 pKa = 6.33SKK73 pKa = 10.98DD74 pKa = 3.56FEE76 pKa = 4.37IKK78 pKa = 10.22SLSKK82 pKa = 10.4KK83 pKa = 10.06IYY85 pKa = 9.47IGVTNTYY92 pKa = 8.16TFEE95 pKa = 4.26AKK97 pKa = 10.24AVFPEE102 pKa = 4.47DD103 pKa = 3.94DD104 pKa = 3.67EE105 pKa = 4.94SLIEE109 pKa = 4.11INFGFYY115 pKa = 10.95YY116 pKa = 10.76LINLYY121 pKa = 10.78GDD123 pKa = 3.85VITRR127 pKa = 11.84LGLTIRR133 pKa = 11.84ADD135 pKa = 4.06FNEE138 pKa = 4.23SSQVLDD144 pKa = 4.85VIDD147 pKa = 4.71SDD149 pKa = 3.64IQKK152 pKa = 10.37GINAIISNFNYY163 pKa = 9.13FQKK166 pKa = 10.71KK167 pKa = 8.79IVSLEE172 pKa = 3.84KK173 pKa = 11.07GPDD176 pKa = 3.24FEE178 pKa = 5.6YY179 pKa = 10.16WGNDD183 pKa = 2.63NWNEE187 pKa = 3.81RR188 pKa = 11.84MEE190 pKa = 4.43NMAFRR195 pKa = 11.84IKK197 pKa = 10.5SIMYY201 pKa = 9.33LFVLLHH207 pKa = 5.9EE208 pKa = 5.06VGHH211 pKa = 6.86HH212 pKa = 6.65IYY214 pKa = 10.98DD215 pKa = 3.65HH216 pKa = 6.76LKK218 pKa = 10.15GFQSYY223 pKa = 10.67SLPDD227 pKa = 3.47IPNQILASIDD237 pKa = 3.45QNKK240 pKa = 9.78YY241 pKa = 10.95NIDD244 pKa = 3.55QIQEE248 pKa = 3.92FEE250 pKa = 4.41ADD252 pKa = 4.04LFAANYY258 pKa = 9.36LLNIRR263 pKa = 11.84DD264 pKa = 3.85YY265 pKa = 11.3TDD267 pKa = 4.37DD268 pKa = 4.97DD269 pKa = 5.23SILKK273 pKa = 10.53LLDD276 pKa = 3.33NTTCSLGILITLHH289 pKa = 5.69LTAGVLGDD297 pKa = 3.71IEE299 pKa = 4.66IGNEE303 pKa = 3.75NHH305 pKa = 7.0PPLSDD310 pKa = 4.49RR311 pKa = 11.84IQIIILFIQQKK322 pKa = 9.65VGQSEE327 pKa = 4.18FNEE330 pKa = 4.8IIDD333 pKa = 3.76TFQNLVYY340 pKa = 10.55RR341 pKa = 11.84IVNFSS346 pKa = 3.0
Molecular weight: 40.2 kDa
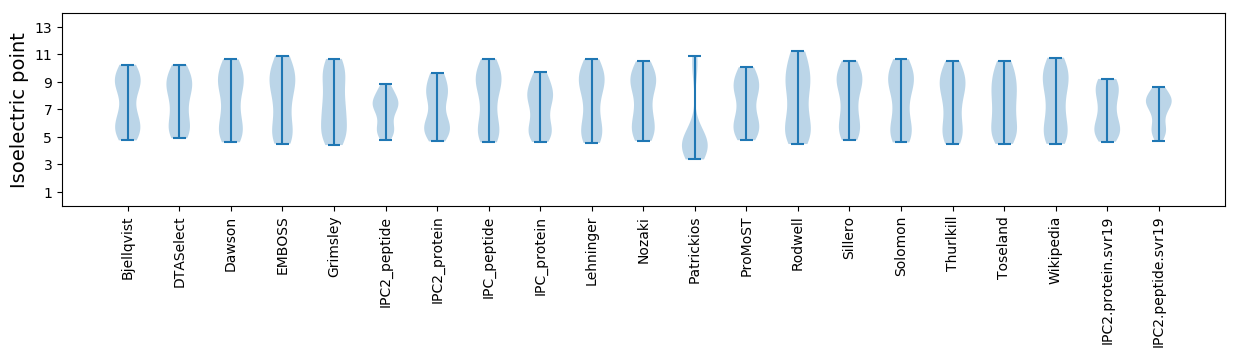
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5WIY1|S5WIY1_9VIRU Uncharacterized protein OS=Leptospira phage vb_LkmZ_Bejolso9-LE1 OX=1334244 GN=LEP1GSC052_0081 PE=4 SV=1
MM1 pKa = 7.56EE2 pKa = 5.27NYY4 pKa = 9.94EE5 pKa = 4.32KK6 pKa = 10.26IKK8 pKa = 10.51RR9 pKa = 11.84LYY11 pKa = 8.44PPYY14 pKa = 9.67WVSDD18 pKa = 3.47VCKK21 pKa = 10.29SLRR24 pKa = 11.84GRR26 pKa = 11.84HH27 pKa = 5.29KK28 pKa = 11.23NIDD31 pKa = 3.22KK32 pKa = 10.7KK33 pKa = 11.1LNSKK37 pKa = 8.23RR38 pKa = 11.84TPKK41 pKa = 9.92PRR43 pKa = 11.84TWKK46 pKa = 10.71LCIEE50 pKa = 4.31YY51 pKa = 10.11NAQKK55 pKa = 10.44LRR57 pKa = 11.84RR58 pKa = 11.84LHH60 pKa = 6.84AKK62 pKa = 10.1DD63 pKa = 3.01GRR65 pKa = 11.84KK66 pKa = 9.56RR67 pKa = 11.84KK68 pKa = 9.57EE69 pKa = 4.01NNQPWNAFVTYY80 pKa = 10.15QKK82 pKa = 10.84EE83 pKa = 3.99KK84 pKa = 10.62LSRR87 pKa = 11.84KK88 pKa = 9.14FDD90 pKa = 3.88EE91 pKa = 4.36NSLWKK96 pKa = 10.36HH97 pKa = 5.47LEE99 pKa = 3.46NRR101 pKa = 11.84ILKK104 pKa = 9.81KK105 pKa = 10.41INAHH109 pKa = 5.58NFPKK113 pKa = 10.41EE114 pKa = 4.03RR115 pKa = 11.84GALWDD120 pKa = 3.57YY121 pKa = 10.74CYY123 pKa = 10.6HH124 pKa = 5.88RR125 pKa = 11.84QIRR128 pKa = 11.84KK129 pKa = 9.15LKK131 pKa = 10.27NIHH134 pKa = 6.57PDD136 pKa = 3.14LRR138 pKa = 11.84EE139 pKa = 3.8RR140 pKa = 11.84PADD143 pKa = 3.59YY144 pKa = 10.53EE145 pKa = 4.0KK146 pKa = 10.99RR147 pKa = 11.84FANRR151 pKa = 11.84QMEE154 pKa = 4.39KK155 pKa = 10.93LKK157 pKa = 10.75FLYY160 pKa = 10.14NKK162 pKa = 9.84KK163 pKa = 10.31KK164 pKa = 10.38PEE166 pKa = 3.83NEE168 pKa = 3.46EE169 pKa = 3.84WKK171 pKa = 10.37FWWLAQLRR179 pKa = 11.84KK180 pKa = 9.8LHH182 pKa = 6.09ARR184 pKa = 11.84YY185 pKa = 9.87KK186 pKa = 10.86GVLGVNNKK194 pKa = 8.64NRR196 pKa = 11.84FKK198 pKa = 11.22DD199 pKa = 3.56NPKK202 pKa = 9.66WYY204 pKa = 10.45
MM1 pKa = 7.56EE2 pKa = 5.27NYY4 pKa = 9.94EE5 pKa = 4.32KK6 pKa = 10.26IKK8 pKa = 10.51RR9 pKa = 11.84LYY11 pKa = 8.44PPYY14 pKa = 9.67WVSDD18 pKa = 3.47VCKK21 pKa = 10.29SLRR24 pKa = 11.84GRR26 pKa = 11.84HH27 pKa = 5.29KK28 pKa = 11.23NIDD31 pKa = 3.22KK32 pKa = 10.7KK33 pKa = 11.1LNSKK37 pKa = 8.23RR38 pKa = 11.84TPKK41 pKa = 9.92PRR43 pKa = 11.84TWKK46 pKa = 10.71LCIEE50 pKa = 4.31YY51 pKa = 10.11NAQKK55 pKa = 10.44LRR57 pKa = 11.84RR58 pKa = 11.84LHH60 pKa = 6.84AKK62 pKa = 10.1DD63 pKa = 3.01GRR65 pKa = 11.84KK66 pKa = 9.56RR67 pKa = 11.84KK68 pKa = 9.57EE69 pKa = 4.01NNQPWNAFVTYY80 pKa = 10.15QKK82 pKa = 10.84EE83 pKa = 3.99KK84 pKa = 10.62LSRR87 pKa = 11.84KK88 pKa = 9.14FDD90 pKa = 3.88EE91 pKa = 4.36NSLWKK96 pKa = 10.36HH97 pKa = 5.47LEE99 pKa = 3.46NRR101 pKa = 11.84ILKK104 pKa = 9.81KK105 pKa = 10.41INAHH109 pKa = 5.58NFPKK113 pKa = 10.41EE114 pKa = 4.03RR115 pKa = 11.84GALWDD120 pKa = 3.57YY121 pKa = 10.74CYY123 pKa = 10.6HH124 pKa = 5.88RR125 pKa = 11.84QIRR128 pKa = 11.84KK129 pKa = 9.15LKK131 pKa = 10.27NIHH134 pKa = 6.57PDD136 pKa = 3.14LRR138 pKa = 11.84EE139 pKa = 3.8RR140 pKa = 11.84PADD143 pKa = 3.59YY144 pKa = 10.53EE145 pKa = 4.0KK146 pKa = 10.99RR147 pKa = 11.84FANRR151 pKa = 11.84QMEE154 pKa = 4.39KK155 pKa = 10.93LKK157 pKa = 10.75FLYY160 pKa = 10.14NKK162 pKa = 9.84KK163 pKa = 10.31KK164 pKa = 10.38PEE166 pKa = 3.83NEE168 pKa = 3.46EE169 pKa = 3.84WKK171 pKa = 10.37FWWLAQLRR179 pKa = 11.84KK180 pKa = 9.8LHH182 pKa = 6.09ARR184 pKa = 11.84YY185 pKa = 9.87KK186 pKa = 10.86GVLGVNNKK194 pKa = 8.64NRR196 pKa = 11.84FKK198 pKa = 11.22DD199 pKa = 3.56NPKK202 pKa = 9.66WYY204 pKa = 10.45
Molecular weight: 25.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13158 |
42 |
2260 |
387.0 |
44.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.264 ± 0.225 | 0.973 ± 0.12 |
5.259 ± 0.292 | 6.407 ± 0.624 |
5.578 ± 0.234 | 5.495 ± 0.672 |
1.695 ± 0.203 | 8.147 ± 0.425 |
8.094 ± 0.563 | 9.941 ± 0.534 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.497 ± 0.08 | 6.954 ± 0.372 |
3.481 ± 0.261 | 3.595 ± 0.162 |
4.309 ± 0.281 | 8.649 ± 0.451 |
5.525 ± 0.598 | 4.651 ± 0.254 |
1.094 ± 0.159 | 4.393 ± 0.168 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
