
Sphingosinithalassobacter sp. zrk23
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingosinithalassobacter; unclassified Sphingosinithalassobacter
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
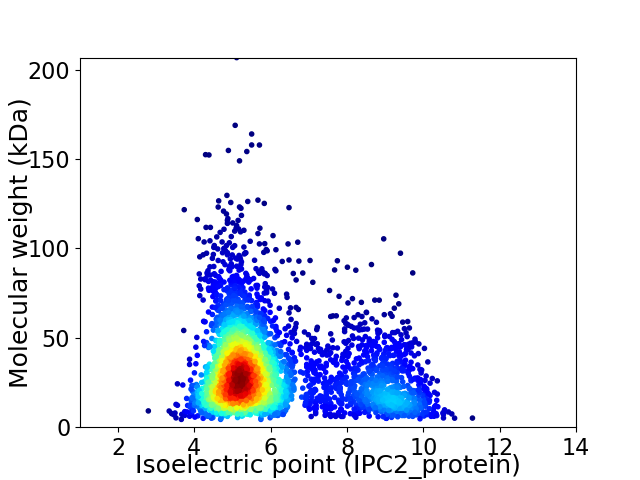
Virtual 2D-PAGE plot for 3726 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6Y8L0|A0A6G6Y8L0_9SPHN Type IV secretion system protein TraC OS=Sphingosinithalassobacter sp. zrk23 OX=2711215 GN=traC PE=4 SV=1
MM1 pKa = 7.6ASIKK5 pKa = 10.21RR6 pKa = 11.84LSLILMTTCAGAALSACDD24 pKa = 3.8GASDD28 pKa = 3.59IASPGTGGNVIINNPAPTPTPTPTPTPTASLVTPATGCPTIADD71 pKa = 4.04PQGLTDD77 pKa = 5.31DD78 pKa = 4.27GTITGPTGEE87 pKa = 4.42YY88 pKa = 9.49RR89 pKa = 11.84VCTLPDD95 pKa = 4.81RR96 pKa = 11.84INTSINLPQIDD107 pKa = 3.99GLLYY111 pKa = 10.46RR112 pKa = 11.84LGGRR116 pKa = 11.84VDD118 pKa = 3.2VGTDD122 pKa = 3.08QGATSTGTDD131 pKa = 3.08VTLSIEE137 pKa = 3.97PGVIIFGGTGVSWLAVNRR155 pKa = 11.84GNAIDD160 pKa = 4.62AVGTPTDD167 pKa = 4.55PIIFTSRR174 pKa = 11.84DD175 pKa = 3.21NVLGLNTDD183 pKa = 3.28SSSGQWGGVVLMGRR197 pKa = 11.84APITDD202 pKa = 3.72CTVTPGATPGTAACEE217 pKa = 4.09RR218 pKa = 11.84QTEE221 pKa = 4.47GAVDD225 pKa = 3.41PAVYY229 pKa = 10.27GGANTADD236 pKa = 3.41SSGTMQYY243 pKa = 10.23VQIRR247 pKa = 11.84YY248 pKa = 9.15SGYY251 pKa = 9.5VLSANNEE258 pKa = 4.15LQSLTTEE265 pKa = 5.03GIGTGTTLDD274 pKa = 4.38HH275 pKa = 6.82IMSFNSSDD283 pKa = 3.69DD284 pKa = 3.56AVEE287 pKa = 4.07MFGGRR292 pKa = 11.84VNMKK296 pKa = 9.72YY297 pKa = 9.73FVAVGAEE304 pKa = 4.14DD305 pKa = 5.43DD306 pKa = 4.99NLDD309 pKa = 3.31TDD311 pKa = 4.16TGVKK315 pKa = 10.07AQLQFVIAAQRR326 pKa = 11.84NGVGDD331 pKa = 3.79TMLEE335 pKa = 3.94ADD337 pKa = 4.56SDD339 pKa = 4.12NSVDD343 pKa = 3.71GDD345 pKa = 3.97LPRR348 pKa = 11.84QHH350 pKa = 6.08TQVANFTFLQRR361 pKa = 11.84SNVGSDD367 pKa = 3.36LAAMYY372 pKa = 10.3IRR374 pKa = 11.84GGADD378 pKa = 2.94YY379 pKa = 11.62SMANGVVSSPNYY391 pKa = 7.59TCLRR395 pKa = 11.84LSRR398 pKa = 11.84TQTIAAANAAIDD410 pKa = 3.95EE411 pKa = 4.52NGPPEE416 pKa = 4.25FEE418 pKa = 4.35SVVMDD423 pKa = 4.39CGTPTFAGNSGVTAADD439 pKa = 3.5VEE441 pKa = 5.17TVFNAGTNNDD451 pKa = 3.88DD452 pKa = 5.52DD453 pKa = 4.5FTTSLTDD460 pKa = 3.22MFVNGSNEE468 pKa = 3.97AGVTAFDD475 pKa = 4.05LTTVGIFWTDD485 pKa = 2.37TDD487 pKa = 4.29YY488 pKa = 11.45IGAVEE493 pKa = 4.75DD494 pKa = 5.98ANDD497 pKa = 3.64TWWQGWTCNSSTLAFAGSSDD517 pKa = 4.12CTSLPTSS524 pKa = 3.58
MM1 pKa = 7.6ASIKK5 pKa = 10.21RR6 pKa = 11.84LSLILMTTCAGAALSACDD24 pKa = 3.8GASDD28 pKa = 3.59IASPGTGGNVIINNPAPTPTPTPTPTPTASLVTPATGCPTIADD71 pKa = 4.04PQGLTDD77 pKa = 5.31DD78 pKa = 4.27GTITGPTGEE87 pKa = 4.42YY88 pKa = 9.49RR89 pKa = 11.84VCTLPDD95 pKa = 4.81RR96 pKa = 11.84INTSINLPQIDD107 pKa = 3.99GLLYY111 pKa = 10.46RR112 pKa = 11.84LGGRR116 pKa = 11.84VDD118 pKa = 3.2VGTDD122 pKa = 3.08QGATSTGTDD131 pKa = 3.08VTLSIEE137 pKa = 3.97PGVIIFGGTGVSWLAVNRR155 pKa = 11.84GNAIDD160 pKa = 4.62AVGTPTDD167 pKa = 4.55PIIFTSRR174 pKa = 11.84DD175 pKa = 3.21NVLGLNTDD183 pKa = 3.28SSSGQWGGVVLMGRR197 pKa = 11.84APITDD202 pKa = 3.72CTVTPGATPGTAACEE217 pKa = 4.09RR218 pKa = 11.84QTEE221 pKa = 4.47GAVDD225 pKa = 3.41PAVYY229 pKa = 10.27GGANTADD236 pKa = 3.41SSGTMQYY243 pKa = 10.23VQIRR247 pKa = 11.84YY248 pKa = 9.15SGYY251 pKa = 9.5VLSANNEE258 pKa = 4.15LQSLTTEE265 pKa = 5.03GIGTGTTLDD274 pKa = 4.38HH275 pKa = 6.82IMSFNSSDD283 pKa = 3.69DD284 pKa = 3.56AVEE287 pKa = 4.07MFGGRR292 pKa = 11.84VNMKK296 pKa = 9.72YY297 pKa = 9.73FVAVGAEE304 pKa = 4.14DD305 pKa = 5.43DD306 pKa = 4.99NLDD309 pKa = 3.31TDD311 pKa = 4.16TGVKK315 pKa = 10.07AQLQFVIAAQRR326 pKa = 11.84NGVGDD331 pKa = 3.79TMLEE335 pKa = 3.94ADD337 pKa = 4.56SDD339 pKa = 4.12NSVDD343 pKa = 3.71GDD345 pKa = 3.97LPRR348 pKa = 11.84QHH350 pKa = 6.08TQVANFTFLQRR361 pKa = 11.84SNVGSDD367 pKa = 3.36LAAMYY372 pKa = 10.3IRR374 pKa = 11.84GGADD378 pKa = 2.94YY379 pKa = 11.62SMANGVVSSPNYY391 pKa = 7.59TCLRR395 pKa = 11.84LSRR398 pKa = 11.84TQTIAAANAAIDD410 pKa = 3.95EE411 pKa = 4.52NGPPEE416 pKa = 4.25FEE418 pKa = 4.35SVVMDD423 pKa = 4.39CGTPTFAGNSGVTAADD439 pKa = 3.5VEE441 pKa = 5.17TVFNAGTNNDD451 pKa = 3.88DD452 pKa = 5.52DD453 pKa = 4.5FTTSLTDD460 pKa = 3.22MFVNGSNEE468 pKa = 3.97AGVTAFDD475 pKa = 4.05LTTVGIFWTDD485 pKa = 2.37TDD487 pKa = 4.29YY488 pKa = 11.45IGAVEE493 pKa = 4.75DD494 pKa = 5.98ANDD497 pKa = 3.64TWWQGWTCNSSTLAFAGSSDD517 pKa = 4.12CTSLPTSS524 pKa = 3.58
Molecular weight: 54.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6Y4E2|A0A6G6Y4E2_9SPHN L D-transpeptidase family protein OS=Sphingosinithalassobacter sp. zrk23 OX=2711215 GN=G5C33_08180 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 9.14VIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 9.14VIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1161024 |
38 |
1918 |
311.6 |
33.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.12 ± 0.06 | 0.785 ± 0.011 |
6.179 ± 0.031 | 6.041 ± 0.037 |
3.557 ± 0.024 | 8.912 ± 0.037 |
1.964 ± 0.019 | 4.963 ± 0.025 |
2.71 ± 0.032 | 9.714 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.512 ± 0.021 | 2.41 ± 0.024 |
5.354 ± 0.03 | 3.055 ± 0.022 |
7.463 ± 0.043 | 5.265 ± 0.033 |
5.199 ± 0.032 | 7.079 ± 0.033 |
1.47 ± 0.017 | 2.245 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
