
Microvirga guangxiensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Microvirga
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
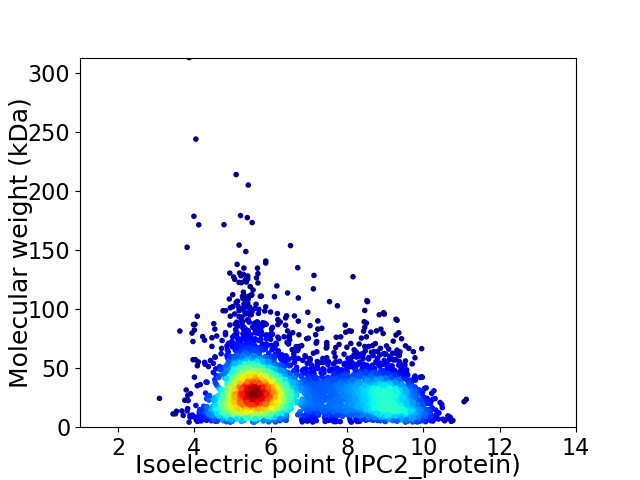
Virtual 2D-PAGE plot for 4496 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G5BFI3|A0A1G5BFI3_9RHIZ Guanylate kinase OS=Microvirga guangxiensis OX=549386 GN=gmk PE=3 SV=1
MM1 pKa = 7.23SRR3 pKa = 11.84KK4 pKa = 9.1EE5 pKa = 3.9NPFNYY10 pKa = 8.52HH11 pKa = 5.96TDD13 pKa = 3.25LSYY16 pKa = 11.48AGLMALQSEE25 pKa = 5.28MIASTLSSAVDD36 pKa = 3.41SYY38 pKa = 11.53FSVDD42 pKa = 2.8RR43 pKa = 11.84GIYY46 pKa = 9.6GGARR50 pKa = 11.84AVWGADD56 pKa = 3.07QVVGCGVDD64 pKa = 3.1WASLYY69 pKa = 10.35PADD72 pKa = 4.49EE73 pKa = 4.27VLDD76 pKa = 4.22GGGGGPGGGPVNSNLPLSCEE96 pKa = 3.97FPPEE100 pKa = 4.19GEE102 pKa = 3.8TRR104 pKa = 11.84IIIEE108 pKa = 4.25STGLPNHH115 pKa = 5.8NRR117 pKa = 11.84VIEE120 pKa = 3.88FHH122 pKa = 6.6TNGTVLIYY130 pKa = 10.21YY131 pKa = 8.3TDD133 pKa = 3.19NDD135 pKa = 3.78GNIRR139 pKa = 11.84GNQYY143 pKa = 10.88SGDD146 pKa = 3.81VQISISTTDD155 pKa = 3.3QPNLFVRR162 pKa = 11.84LEE164 pKa = 4.07EE165 pKa = 4.38YY166 pKa = 10.33PDD168 pKa = 4.18GSWVEE173 pKa = 4.21SIIDD177 pKa = 3.16IDD179 pKa = 4.35GNVLEE184 pKa = 4.63WEE186 pKa = 4.58TYY188 pKa = 8.89TVVDD192 pKa = 4.17NPDD195 pKa = 3.01GTRR198 pKa = 11.84TDD200 pKa = 2.92IYY202 pKa = 10.83YY203 pKa = 10.5DD204 pKa = 3.88SNGHH208 pKa = 5.62GSRR211 pKa = 11.84LTYY214 pKa = 10.78SSDD217 pKa = 3.87DD218 pKa = 3.46VSPWSEE224 pKa = 4.4TYY226 pKa = 10.27PDD228 pKa = 3.68DD229 pKa = 5.16AIRR232 pKa = 11.84PISSVPPPEE241 pKa = 5.23GEE243 pKa = 3.83TRR245 pKa = 11.84TVIEE249 pKa = 4.17STGVSGHH256 pKa = 5.14NRR258 pKa = 11.84VVEE261 pKa = 5.12FYY263 pKa = 11.47SNGTVLIYY271 pKa = 10.22YY272 pKa = 8.4TDD274 pKa = 3.06NDD276 pKa = 3.72GNLRR280 pKa = 11.84GNQYY284 pKa = 10.92SGDD287 pKa = 3.49VKK289 pKa = 10.32VTIKK293 pKa = 9.61EE294 pKa = 4.32TEE296 pKa = 4.0EE297 pKa = 3.53PNIYY301 pKa = 10.36YY302 pKa = 10.06RR303 pKa = 11.84VEE305 pKa = 4.1EE306 pKa = 4.35YY307 pKa = 10.3PDD309 pKa = 4.11GSWVEE314 pKa = 4.04TIIDD318 pKa = 3.16IDD320 pKa = 4.07ANVIGWEE327 pKa = 4.03SSTIIDD333 pKa = 4.31NPDD336 pKa = 3.01GTKK339 pKa = 9.98TEE341 pKa = 3.97IYY343 pKa = 8.93RR344 pKa = 11.84DD345 pKa = 3.46SMGKK349 pKa = 8.17MRR351 pKa = 11.84LTSGQSTVQYY361 pKa = 10.76GKK363 pKa = 10.92VGDD366 pKa = 3.84DD367 pKa = 3.54HH368 pKa = 8.13LLGGSGNDD376 pKa = 3.29RR377 pKa = 11.84LKK379 pKa = 11.05GASGQDD385 pKa = 3.49TLDD388 pKa = 3.47GSAGDD393 pKa = 3.85DD394 pKa = 3.51ALFGEE399 pKa = 5.17AGDD402 pKa = 4.56DD403 pKa = 3.87LLDD406 pKa = 4.35GGVGDD411 pKa = 5.28DD412 pKa = 4.43LLLGGVGNDD421 pKa = 3.41LALGGEE427 pKa = 4.55GRR429 pKa = 11.84DD430 pKa = 3.88TLDD433 pKa = 3.62GGAGNDD439 pKa = 3.73TLNGGGGFDD448 pKa = 4.39LLSGGSGDD456 pKa = 4.46DD457 pKa = 3.93VISGGSGNDD466 pKa = 3.63LLNGQDD472 pKa = 4.69GNDD475 pKa = 3.45ILQGDD480 pKa = 4.49DD481 pKa = 3.55GADD484 pKa = 3.5FLTGGAGDD492 pKa = 3.64DD493 pKa = 4.22TIRR496 pKa = 11.84GGSDD500 pKa = 3.04NDD502 pKa = 3.81SLFGNEE508 pKa = 5.06GNDD511 pKa = 3.86LLDD514 pKa = 4.27GGSGNDD520 pKa = 3.58EE521 pKa = 4.41LYY523 pKa = 11.3GLAGNDD529 pKa = 3.73MLDD532 pKa = 3.49GGAGNDD538 pKa = 4.88LMDD541 pKa = 4.95GDD543 pKa = 4.69EE544 pKa = 4.99GADD547 pKa = 3.58TLLGGAGNDD556 pKa = 3.61YY557 pKa = 10.49MRR559 pKa = 11.84GGVGADD565 pKa = 3.27VLDD568 pKa = 4.29GGEE571 pKa = 4.8GIDD574 pKa = 3.52TVDD577 pKa = 3.94YY578 pKa = 10.61SGSDD582 pKa = 2.97AGIVVNLSTGIGTGGHH598 pKa = 6.16AQDD601 pKa = 3.92DD602 pKa = 4.02VLRR605 pKa = 11.84NIEE608 pKa = 4.1NVIGSAYY615 pKa = 10.33NDD617 pKa = 4.06LLVGHH622 pKa = 7.58AGHH625 pKa = 7.28DD626 pKa = 3.9VLWGNDD632 pKa = 3.57GNDD635 pKa = 4.0TIHH638 pKa = 7.09GGLGNDD644 pKa = 3.94TLHH647 pKa = 7.1GEE649 pKa = 4.3AGDD652 pKa = 3.84DD653 pKa = 3.88HH654 pKa = 6.53MSGGAGHH661 pKa = 7.06DD662 pKa = 3.8TLHH665 pKa = 6.83GGDD668 pKa = 4.69GQDD671 pKa = 3.15QLYY674 pKa = 10.98GEE676 pKa = 5.49DD677 pKa = 4.14GNDD680 pKa = 3.79ALRR683 pKa = 11.84GDD685 pKa = 4.39AGNDD689 pKa = 3.26SLYY692 pKa = 11.21GGAGSDD698 pKa = 3.64EE699 pKa = 5.25LIGGTGDD706 pKa = 3.3DD707 pKa = 4.37HH708 pKa = 7.26LAGGLDD714 pKa = 3.6QDD716 pKa = 3.78WLYY719 pKa = 11.6GGDD722 pKa = 3.9GQDD725 pKa = 2.69WLYY728 pKa = 11.64GEE730 pKa = 5.35DD731 pKa = 4.46GNDD734 pKa = 3.13ILHH737 pKa = 6.07GQNGNDD743 pKa = 3.49YY744 pKa = 10.88LYY746 pKa = 11.04GGNGNEE752 pKa = 4.07LDD754 pKa = 3.64AGGSVFLEE762 pKa = 4.97GIKK765 pKa = 10.47AEE767 pKa = 3.97QLKK770 pKa = 10.99GGIVDD775 pKa = 5.64GAWQWWMM782 pKa = 3.74
MM1 pKa = 7.23SRR3 pKa = 11.84KK4 pKa = 9.1EE5 pKa = 3.9NPFNYY10 pKa = 8.52HH11 pKa = 5.96TDD13 pKa = 3.25LSYY16 pKa = 11.48AGLMALQSEE25 pKa = 5.28MIASTLSSAVDD36 pKa = 3.41SYY38 pKa = 11.53FSVDD42 pKa = 2.8RR43 pKa = 11.84GIYY46 pKa = 9.6GGARR50 pKa = 11.84AVWGADD56 pKa = 3.07QVVGCGVDD64 pKa = 3.1WASLYY69 pKa = 10.35PADD72 pKa = 4.49EE73 pKa = 4.27VLDD76 pKa = 4.22GGGGGPGGGPVNSNLPLSCEE96 pKa = 3.97FPPEE100 pKa = 4.19GEE102 pKa = 3.8TRR104 pKa = 11.84IIIEE108 pKa = 4.25STGLPNHH115 pKa = 5.8NRR117 pKa = 11.84VIEE120 pKa = 3.88FHH122 pKa = 6.6TNGTVLIYY130 pKa = 10.21YY131 pKa = 8.3TDD133 pKa = 3.19NDD135 pKa = 3.78GNIRR139 pKa = 11.84GNQYY143 pKa = 10.88SGDD146 pKa = 3.81VQISISTTDD155 pKa = 3.3QPNLFVRR162 pKa = 11.84LEE164 pKa = 4.07EE165 pKa = 4.38YY166 pKa = 10.33PDD168 pKa = 4.18GSWVEE173 pKa = 4.21SIIDD177 pKa = 3.16IDD179 pKa = 4.35GNVLEE184 pKa = 4.63WEE186 pKa = 4.58TYY188 pKa = 8.89TVVDD192 pKa = 4.17NPDD195 pKa = 3.01GTRR198 pKa = 11.84TDD200 pKa = 2.92IYY202 pKa = 10.83YY203 pKa = 10.5DD204 pKa = 3.88SNGHH208 pKa = 5.62GSRR211 pKa = 11.84LTYY214 pKa = 10.78SSDD217 pKa = 3.87DD218 pKa = 3.46VSPWSEE224 pKa = 4.4TYY226 pKa = 10.27PDD228 pKa = 3.68DD229 pKa = 5.16AIRR232 pKa = 11.84PISSVPPPEE241 pKa = 5.23GEE243 pKa = 3.83TRR245 pKa = 11.84TVIEE249 pKa = 4.17STGVSGHH256 pKa = 5.14NRR258 pKa = 11.84VVEE261 pKa = 5.12FYY263 pKa = 11.47SNGTVLIYY271 pKa = 10.22YY272 pKa = 8.4TDD274 pKa = 3.06NDD276 pKa = 3.72GNLRR280 pKa = 11.84GNQYY284 pKa = 10.92SGDD287 pKa = 3.49VKK289 pKa = 10.32VTIKK293 pKa = 9.61EE294 pKa = 4.32TEE296 pKa = 4.0EE297 pKa = 3.53PNIYY301 pKa = 10.36YY302 pKa = 10.06RR303 pKa = 11.84VEE305 pKa = 4.1EE306 pKa = 4.35YY307 pKa = 10.3PDD309 pKa = 4.11GSWVEE314 pKa = 4.04TIIDD318 pKa = 3.16IDD320 pKa = 4.07ANVIGWEE327 pKa = 4.03SSTIIDD333 pKa = 4.31NPDD336 pKa = 3.01GTKK339 pKa = 9.98TEE341 pKa = 3.97IYY343 pKa = 8.93RR344 pKa = 11.84DD345 pKa = 3.46SMGKK349 pKa = 8.17MRR351 pKa = 11.84LTSGQSTVQYY361 pKa = 10.76GKK363 pKa = 10.92VGDD366 pKa = 3.84DD367 pKa = 3.54HH368 pKa = 8.13LLGGSGNDD376 pKa = 3.29RR377 pKa = 11.84LKK379 pKa = 11.05GASGQDD385 pKa = 3.49TLDD388 pKa = 3.47GSAGDD393 pKa = 3.85DD394 pKa = 3.51ALFGEE399 pKa = 5.17AGDD402 pKa = 4.56DD403 pKa = 3.87LLDD406 pKa = 4.35GGVGDD411 pKa = 5.28DD412 pKa = 4.43LLLGGVGNDD421 pKa = 3.41LALGGEE427 pKa = 4.55GRR429 pKa = 11.84DD430 pKa = 3.88TLDD433 pKa = 3.62GGAGNDD439 pKa = 3.73TLNGGGGFDD448 pKa = 4.39LLSGGSGDD456 pKa = 4.46DD457 pKa = 3.93VISGGSGNDD466 pKa = 3.63LLNGQDD472 pKa = 4.69GNDD475 pKa = 3.45ILQGDD480 pKa = 4.49DD481 pKa = 3.55GADD484 pKa = 3.5FLTGGAGDD492 pKa = 3.64DD493 pKa = 4.22TIRR496 pKa = 11.84GGSDD500 pKa = 3.04NDD502 pKa = 3.81SLFGNEE508 pKa = 5.06GNDD511 pKa = 3.86LLDD514 pKa = 4.27GGSGNDD520 pKa = 3.58EE521 pKa = 4.41LYY523 pKa = 11.3GLAGNDD529 pKa = 3.73MLDD532 pKa = 3.49GGAGNDD538 pKa = 4.88LMDD541 pKa = 4.95GDD543 pKa = 4.69EE544 pKa = 4.99GADD547 pKa = 3.58TLLGGAGNDD556 pKa = 3.61YY557 pKa = 10.49MRR559 pKa = 11.84GGVGADD565 pKa = 3.27VLDD568 pKa = 4.29GGEE571 pKa = 4.8GIDD574 pKa = 3.52TVDD577 pKa = 3.94YY578 pKa = 10.61SGSDD582 pKa = 2.97AGIVVNLSTGIGTGGHH598 pKa = 6.16AQDD601 pKa = 3.92DD602 pKa = 4.02VLRR605 pKa = 11.84NIEE608 pKa = 4.1NVIGSAYY615 pKa = 10.33NDD617 pKa = 4.06LLVGHH622 pKa = 7.58AGHH625 pKa = 7.28DD626 pKa = 3.9VLWGNDD632 pKa = 3.57GNDD635 pKa = 4.0TIHH638 pKa = 7.09GGLGNDD644 pKa = 3.94TLHH647 pKa = 7.1GEE649 pKa = 4.3AGDD652 pKa = 3.84DD653 pKa = 3.88HH654 pKa = 6.53MSGGAGHH661 pKa = 7.06DD662 pKa = 3.8TLHH665 pKa = 6.83GGDD668 pKa = 4.69GQDD671 pKa = 3.15QLYY674 pKa = 10.98GEE676 pKa = 5.49DD677 pKa = 4.14GNDD680 pKa = 3.79ALRR683 pKa = 11.84GDD685 pKa = 4.39AGNDD689 pKa = 3.26SLYY692 pKa = 11.21GGAGSDD698 pKa = 3.64EE699 pKa = 5.25LIGGTGDD706 pKa = 3.3DD707 pKa = 4.37HH708 pKa = 7.26LAGGLDD714 pKa = 3.6QDD716 pKa = 3.78WLYY719 pKa = 11.6GGDD722 pKa = 3.9GQDD725 pKa = 2.69WLYY728 pKa = 11.64GEE730 pKa = 5.35DD731 pKa = 4.46GNDD734 pKa = 3.13ILHH737 pKa = 6.07GQNGNDD743 pKa = 3.49YY744 pKa = 10.88LYY746 pKa = 11.04GGNGNEE752 pKa = 4.07LDD754 pKa = 3.64AGGSVFLEE762 pKa = 4.97GIKK765 pKa = 10.47AEE767 pKa = 3.97QLKK770 pKa = 10.99GGIVDD775 pKa = 5.64GAWQWWMM782 pKa = 3.74
Molecular weight: 81.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G5JKX3|A0A1G5JKX3_9RHIZ Predicted Zn-dependent peptidase OS=Microvirga guangxiensis OX=549386 GN=SAMN02927923_02713 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.23KK3 pKa = 9.93ARR5 pKa = 11.84PVSRR9 pKa = 11.84VARR12 pKa = 11.84AVPGAARR19 pKa = 11.84KK20 pKa = 9.13VVSGPSVAVAVPAEE34 pKa = 3.74ARR36 pKa = 11.84LRR38 pKa = 11.84HH39 pKa = 5.87PVQPPARR46 pKa = 11.84ARR48 pKa = 11.84SGSHH52 pKa = 5.46LRR54 pKa = 11.84RR55 pKa = 11.84HH56 pKa = 5.64RR57 pKa = 11.84VLLKK61 pKa = 10.55SNVRR65 pKa = 11.84NRR67 pKa = 11.84GEE69 pKa = 4.01NSASRR74 pKa = 11.84QNALPRR80 pKa = 11.84QLLPSRR86 pKa = 11.84GQPLRR91 pKa = 11.84NEE93 pKa = 3.57LRR95 pKa = 11.84TRR97 pKa = 11.84HH98 pKa = 5.74LRR100 pKa = 11.84GLSAQTLRR108 pKa = 11.84SVSVPPAPMHH118 pKa = 6.62RR119 pKa = 11.84SANSVPQSASALRR132 pKa = 11.84LPMPRR137 pKa = 11.84RR138 pKa = 11.84GLRR141 pKa = 11.84RR142 pKa = 11.84PLLLNRR148 pKa = 11.84PRR150 pKa = 11.84RR151 pKa = 11.84LPRR154 pKa = 11.84QSLEE158 pKa = 3.62RR159 pKa = 11.84HH160 pKa = 5.73RR161 pKa = 11.84PRR163 pKa = 11.84LAPAMRR169 pKa = 11.84NVPIKK174 pKa = 10.52VSRR177 pKa = 11.84GPLRR181 pKa = 11.84HH182 pKa = 6.41LPPPSRR188 pKa = 11.84SGAHH192 pKa = 5.3RR193 pKa = 11.84QQ194 pKa = 3.23
MM1 pKa = 7.44KK2 pKa = 10.23KK3 pKa = 9.93ARR5 pKa = 11.84PVSRR9 pKa = 11.84VARR12 pKa = 11.84AVPGAARR19 pKa = 11.84KK20 pKa = 9.13VVSGPSVAVAVPAEE34 pKa = 3.74ARR36 pKa = 11.84LRR38 pKa = 11.84HH39 pKa = 5.87PVQPPARR46 pKa = 11.84ARR48 pKa = 11.84SGSHH52 pKa = 5.46LRR54 pKa = 11.84RR55 pKa = 11.84HH56 pKa = 5.64RR57 pKa = 11.84VLLKK61 pKa = 10.55SNVRR65 pKa = 11.84NRR67 pKa = 11.84GEE69 pKa = 4.01NSASRR74 pKa = 11.84QNALPRR80 pKa = 11.84QLLPSRR86 pKa = 11.84GQPLRR91 pKa = 11.84NEE93 pKa = 3.57LRR95 pKa = 11.84TRR97 pKa = 11.84HH98 pKa = 5.74LRR100 pKa = 11.84GLSAQTLRR108 pKa = 11.84SVSVPPAPMHH118 pKa = 6.62RR119 pKa = 11.84SANSVPQSASALRR132 pKa = 11.84LPMPRR137 pKa = 11.84RR138 pKa = 11.84GLRR141 pKa = 11.84RR142 pKa = 11.84PLLLNRR148 pKa = 11.84PRR150 pKa = 11.84RR151 pKa = 11.84LPRR154 pKa = 11.84QSLEE158 pKa = 3.62RR159 pKa = 11.84HH160 pKa = 5.73RR161 pKa = 11.84PRR163 pKa = 11.84LAPAMRR169 pKa = 11.84NVPIKK174 pKa = 10.52VSRR177 pKa = 11.84GPLRR181 pKa = 11.84HH182 pKa = 6.41LPPPSRR188 pKa = 11.84SGAHH192 pKa = 5.3RR193 pKa = 11.84QQ194 pKa = 3.23
Molecular weight: 21.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1369810 |
39 |
3014 |
304.7 |
33.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.654 ± 0.046 | 0.781 ± 0.011 |
5.408 ± 0.032 | 5.978 ± 0.036 |
3.724 ± 0.023 | 8.406 ± 0.043 |
2.018 ± 0.019 | 5.474 ± 0.027 |
3.552 ± 0.029 | 10.219 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.484 ± 0.015 | 2.732 ± 0.026 |
5.162 ± 0.033 | 3.269 ± 0.019 |
7.03 ± 0.039 | 5.788 ± 0.027 |
5.324 ± 0.03 | 7.459 ± 0.027 |
1.297 ± 0.016 | 2.241 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
