
Chino del tomate Amazonas virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.57
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|E9KZK8|E9KZK8_9GEMI Replication-associated protein OS=Chino del tomate Amazonas virus OX=858516 GN=AC1 PE=3 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84SSSPSAPPSIKK13 pKa = 9.69RR14 pKa = 11.84AHH16 pKa = 5.99RR17 pKa = 11.84QAKK20 pKa = 8.85KK21 pKa = 9.0RR22 pKa = 11.84AIRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84IDD30 pKa = 3.78LEE32 pKa = 4.52CGCSIYY38 pKa = 11.2LHH40 pKa = 6.75IGCTGHH46 pKa = 6.92GFTHH50 pKa = 7.49RR51 pKa = 11.84GTHH54 pKa = 5.78HH55 pKa = 6.16CTSGRR60 pKa = 11.84EE61 pKa = 3.81WRR63 pKa = 11.84VYY65 pKa = 10.84LGDD68 pKa = 3.55NKK70 pKa = 10.88SPIFQDD76 pKa = 2.83IQRR79 pKa = 11.84RR80 pKa = 11.84DD81 pKa = 3.48THH83 pKa = 5.04VHH85 pKa = 5.47EE86 pKa = 4.69LQGVSHH92 pKa = 7.06TNTVQSQPQEE102 pKa = 4.3GVEE105 pKa = 4.43STQSHH110 pKa = 7.1PEE112 pKa = 3.94FPSLDD117 pKa = 4.61DD118 pKa = 4.15ISDD121 pKa = 4.01SFWDD125 pKa = 5.14DD126 pKa = 2.62IFKK129 pKa = 11.03
MM1 pKa = 7.7RR2 pKa = 11.84SSSPSAPPSIKK13 pKa = 9.69RR14 pKa = 11.84AHH16 pKa = 5.99RR17 pKa = 11.84QAKK20 pKa = 8.85KK21 pKa = 9.0RR22 pKa = 11.84AIRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84IDD30 pKa = 3.78LEE32 pKa = 4.52CGCSIYY38 pKa = 11.2LHH40 pKa = 6.75IGCTGHH46 pKa = 6.92GFTHH50 pKa = 7.49RR51 pKa = 11.84GTHH54 pKa = 5.78HH55 pKa = 6.16CTSGRR60 pKa = 11.84EE61 pKa = 3.81WRR63 pKa = 11.84VYY65 pKa = 10.84LGDD68 pKa = 3.55NKK70 pKa = 10.88SPIFQDD76 pKa = 2.83IQRR79 pKa = 11.84RR80 pKa = 11.84DD81 pKa = 3.48THH83 pKa = 5.04VHH85 pKa = 5.47EE86 pKa = 4.69LQGVSHH92 pKa = 7.06TNTVQSQPQEE102 pKa = 4.3GVEE105 pKa = 4.43STQSHH110 pKa = 7.1PEE112 pKa = 3.94FPSLDD117 pKa = 4.61DD118 pKa = 4.15ISDD121 pKa = 4.01SFWDD125 pKa = 5.14DD126 pKa = 2.62IFKK129 pKa = 11.03
Molecular weight: 14.78 kDa
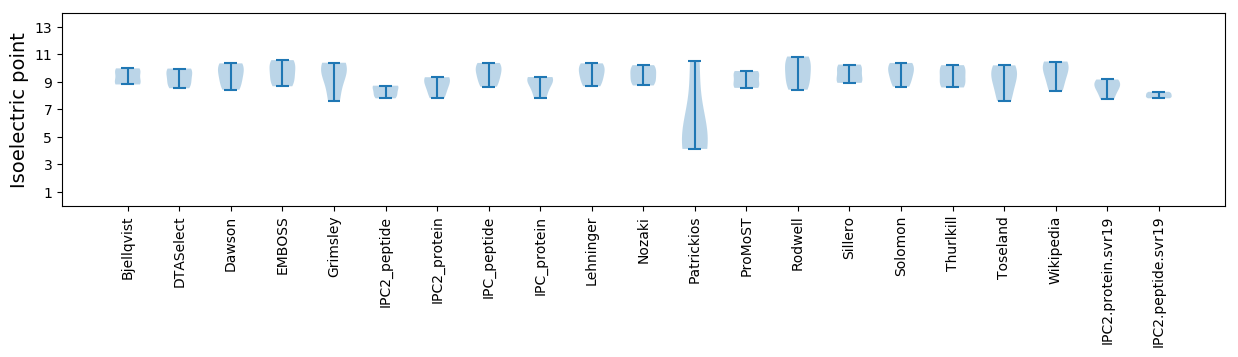
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E9KZK8|E9KZK8_9GEMI Replication-associated protein OS=Chino del tomate Amazonas virus OX=858516 GN=AC1 PE=3 SV=1
MM1 pKa = 7.18SQFSEE6 pKa = 4.14SSNWGDD12 pKa = 3.88KK13 pKa = 10.67PILPKK18 pKa = 10.3CHH20 pKa = 6.59HH21 pKa = 6.82PSVSKK26 pKa = 10.74SLPKK30 pKa = 10.12TISLLILIALSPKK43 pKa = 9.49TMHH46 pKa = 6.61FPNSSTFKK54 pKa = 10.35PQLTNYY60 pKa = 10.16SFVLQEE66 pKa = 3.78SFTRR70 pKa = 11.84MGSLISMCLSNSKK83 pKa = 10.92GNTNAPTTDD92 pKa = 2.73SSTWYY97 pKa = 8.26PQPDD101 pKa = 3.2QHH103 pKa = 8.64ISIRR107 pKa = 11.84TFRR110 pKa = 11.84EE111 pKa = 3.78LKK113 pKa = 10.48KK114 pKa = 10.21QAQTSKK120 pKa = 10.42PIWRR124 pKa = 11.84KK125 pKa = 7.83TEE127 pKa = 3.48TSLIMEE133 pKa = 5.58FSRR136 pKa = 11.84SMDD139 pKa = 3.52DD140 pKa = 3.58QLEE143 pKa = 4.27EE144 pKa = 4.17VANLPTTLMPRR155 pKa = 11.84LSMQAMQPRR164 pKa = 11.84PSIYY168 pKa = 9.77SEE170 pKa = 4.02RR171 pKa = 11.84NNPEE175 pKa = 3.5TFLQIIIISKK185 pKa = 9.53QMQKK189 pKa = 10.79GFLLRR194 pKa = 11.84LRR196 pKa = 11.84NHH198 pKa = 6.42GFLRR202 pKa = 11.84FPSPHH207 pKa = 6.65SLTFQKK213 pKa = 10.28RR214 pKa = 11.84CKK216 pKa = 9.76SGQIIISGEE225 pKa = 3.83VPLRR229 pKa = 11.84GRR231 pKa = 11.84RR232 pKa = 11.84DD233 pKa = 3.43RR234 pKa = 11.84LVSSSRR240 pKa = 11.84VNSRR244 pKa = 11.84TGKK247 pKa = 8.38TMWARR252 pKa = 11.84ALGPHH257 pKa = 6.86TYY259 pKa = 10.87LSGHH263 pKa = 7.06LDD265 pKa = 3.61FNGTVYY271 pKa = 10.85SNQVEE276 pKa = 4.58YY277 pKa = 10.36TVIAAVAPHH286 pKa = 5.5YY287 pKa = 10.87LKK289 pKa = 10.79LKK291 pKa = 9.12HH292 pKa = 5.78WKK294 pKa = 9.68EE295 pKa = 4.26LIGSQKK301 pKa = 10.68DD302 pKa = 3.23WQSNCKK308 pKa = 9.38YY309 pKa = 9.74GKK311 pKa = 9.18PVQIQGGIPSIVLCNPGSGFQLSRR335 pKa = 11.84FPQQRR340 pKa = 11.84GKK342 pKa = 11.01RR343 pKa = 11.84ITQEE347 pKa = 4.33LDD349 pKa = 3.19PQEE352 pKa = 4.86CALHH356 pKa = 6.66HH357 pKa = 6.4PQLPPQQ363 pKa = 3.65
MM1 pKa = 7.18SQFSEE6 pKa = 4.14SSNWGDD12 pKa = 3.88KK13 pKa = 10.67PILPKK18 pKa = 10.3CHH20 pKa = 6.59HH21 pKa = 6.82PSVSKK26 pKa = 10.74SLPKK30 pKa = 10.12TISLLILIALSPKK43 pKa = 9.49TMHH46 pKa = 6.61FPNSSTFKK54 pKa = 10.35PQLTNYY60 pKa = 10.16SFVLQEE66 pKa = 3.78SFTRR70 pKa = 11.84MGSLISMCLSNSKK83 pKa = 10.92GNTNAPTTDD92 pKa = 2.73SSTWYY97 pKa = 8.26PQPDD101 pKa = 3.2QHH103 pKa = 8.64ISIRR107 pKa = 11.84TFRR110 pKa = 11.84EE111 pKa = 3.78LKK113 pKa = 10.48KK114 pKa = 10.21QAQTSKK120 pKa = 10.42PIWRR124 pKa = 11.84KK125 pKa = 7.83TEE127 pKa = 3.48TSLIMEE133 pKa = 5.58FSRR136 pKa = 11.84SMDD139 pKa = 3.52DD140 pKa = 3.58QLEE143 pKa = 4.27EE144 pKa = 4.17VANLPTTLMPRR155 pKa = 11.84LSMQAMQPRR164 pKa = 11.84PSIYY168 pKa = 9.77SEE170 pKa = 4.02RR171 pKa = 11.84NNPEE175 pKa = 3.5TFLQIIIISKK185 pKa = 9.53QMQKK189 pKa = 10.79GFLLRR194 pKa = 11.84LRR196 pKa = 11.84NHH198 pKa = 6.42GFLRR202 pKa = 11.84FPSPHH207 pKa = 6.65SLTFQKK213 pKa = 10.28RR214 pKa = 11.84CKK216 pKa = 9.76SGQIIISGEE225 pKa = 3.83VPLRR229 pKa = 11.84GRR231 pKa = 11.84RR232 pKa = 11.84DD233 pKa = 3.43RR234 pKa = 11.84LVSSSRR240 pKa = 11.84VNSRR244 pKa = 11.84TGKK247 pKa = 8.38TMWARR252 pKa = 11.84ALGPHH257 pKa = 6.86TYY259 pKa = 10.87LSGHH263 pKa = 7.06LDD265 pKa = 3.61FNGTVYY271 pKa = 10.85SNQVEE276 pKa = 4.58YY277 pKa = 10.36TVIAAVAPHH286 pKa = 5.5YY287 pKa = 10.87LKK289 pKa = 10.79LKK291 pKa = 9.12HH292 pKa = 5.78WKK294 pKa = 9.68EE295 pKa = 4.26LIGSQKK301 pKa = 10.68DD302 pKa = 3.23WQSNCKK308 pKa = 9.38YY309 pKa = 9.74GKK311 pKa = 9.18PVQIQGGIPSIVLCNPGSGFQLSRR335 pKa = 11.84FPQQRR340 pKa = 11.84GKK342 pKa = 11.01RR343 pKa = 11.84ITQEE347 pKa = 4.33LDD349 pKa = 3.19PQEE352 pKa = 4.86CALHH356 pKa = 6.66HH357 pKa = 6.4PQLPPQQ363 pKa = 3.65
Molecular weight: 41.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
883 |
125 |
363 |
220.8 |
25.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.077 ± 0.689 | 1.812 ± 0.388 |
3.398 ± 0.688 | 4.19 ± 0.342 |
4.077 ± 0.047 | 5.776 ± 0.81 |
4.19 ± 0.744 | 6.455 ± 0.388 |
5.436 ± 0.559 | 7.701 ± 1.14 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.945 ± 0.514 | 5.21 ± 1.155 |
5.663 ± 1.353 | 5.323 ± 1.264 |
7.135 ± 0.704 | 9.626 ± 1.484 |
6.455 ± 0.666 | 5.436 ± 1.332 |
1.586 ± 0.038 | 3.511 ± 0.952 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
