
Halorubrum pleomorphic virus 2
Taxonomy: Viruses; Monodnaviria; Trapavirae; Saleviricota; Huolimaviricetes; Haloruvirales; Pleolipoviridae; Alphapleolipovirus; Alphapleolipovirus HRPV2
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
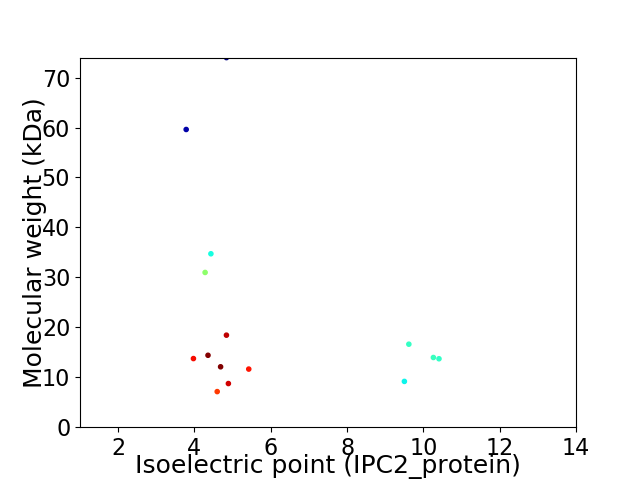
Virtual 2D-PAGE plot for 15 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H9ABM0|H9ABM0_9VIRU ORF6 OS=Halorubrum pleomorphic virus 2 OX=1156719 GN=ORF6 PE=4 SV=1
MM1 pKa = 7.55GRR3 pKa = 11.84RR4 pKa = 11.84TFVKK8 pKa = 10.51GLGAAGAATVGLSVDD23 pKa = 4.34DD24 pKa = 5.68GFAQDD29 pKa = 3.91AQAIAPLVGVGLAAGAVGVGWALRR53 pKa = 11.84EE54 pKa = 3.93FEE56 pKa = 4.85IVGSDD61 pKa = 3.65APPEE65 pKa = 4.3GLTADD70 pKa = 3.62ALKK73 pKa = 10.31QQVYY77 pKa = 7.21QTAKK81 pKa = 8.92TRR83 pKa = 11.84KK84 pKa = 7.73STNASTIVDD93 pKa = 3.75NQNILDD99 pKa = 4.23GVKK102 pKa = 8.61HH103 pKa = 5.26TAYY106 pKa = 9.75TDD108 pKa = 3.32AKK110 pKa = 9.46IAAIEE115 pKa = 3.95EE116 pKa = 4.46LNAGSAEE123 pKa = 4.21SAVLDD128 pKa = 4.02AATTEE133 pKa = 4.3VNSYY137 pKa = 8.69LTTVQSNFLKK147 pKa = 9.59TWNEE151 pKa = 3.9SVAEE155 pKa = 4.06LDD157 pKa = 4.39SILSTVVNHH166 pKa = 6.72PDD168 pKa = 2.91IGKK171 pKa = 10.49GDD173 pKa = 3.92VFLMLNGSDD182 pKa = 3.54NTIEE186 pKa = 5.01DD187 pKa = 4.15LLANPSGSTDD197 pKa = 3.02ATSFTLADD205 pKa = 3.87GTTMSVGTVEE215 pKa = 4.02VDD217 pKa = 2.94RR218 pKa = 11.84GTEE221 pKa = 3.77SYY223 pKa = 11.09YY224 pKa = 10.34YY225 pKa = 10.72DD226 pKa = 3.98PMSGLVGDD234 pKa = 5.65LGDD237 pKa = 4.7LKK239 pKa = 11.3NGGPTVQYY247 pKa = 10.7DD248 pKa = 3.95GNSLVYY254 pKa = 10.65LNASNWKK261 pKa = 9.34PIYY264 pKa = 10.52DD265 pKa = 3.79EE266 pKa = 4.63MDD268 pKa = 3.1TVLQNVRR275 pKa = 11.84SGISTWVSNVYY286 pKa = 11.0GDD288 pKa = 3.81VQSGEE293 pKa = 4.29IEE295 pKa = 4.27VSDD298 pKa = 3.65LVTPRR303 pKa = 11.84EE304 pKa = 4.0RR305 pKa = 11.84AAMMAQEE312 pKa = 4.65EE313 pKa = 4.97GMSQAIADD321 pKa = 5.12LIALNVPVDD330 pKa = 3.84AEE332 pKa = 4.23RR333 pKa = 11.84EE334 pKa = 4.21ATITIQDD341 pKa = 3.56TGATLPGTFALTDD354 pKa = 3.64ASDD357 pKa = 4.16GPLEE361 pKa = 4.31SGKK364 pKa = 9.08TYY366 pKa = 10.79DD367 pKa = 3.71PSTFSGDD374 pKa = 3.59VYY376 pKa = 9.75FTADD380 pKa = 3.14MSLVEE385 pKa = 4.73GDD387 pKa = 2.89WTAYY391 pKa = 9.88QSGVDD396 pKa = 3.85GGNVTLTSEE405 pKa = 4.71PYY407 pKa = 10.31SGTAVEE413 pKa = 5.43LNTAANEE420 pKa = 4.25TVAVDD425 pKa = 3.73AGNWTATGNGTWYY438 pKa = 10.52HH439 pKa = 6.33DD440 pKa = 3.71VSPEE444 pKa = 4.05LEE446 pKa = 3.93TDD448 pKa = 2.85ITSIEE453 pKa = 4.21SARR456 pKa = 11.84FLSTAEE462 pKa = 3.68QTQYY466 pKa = 9.94EE467 pKa = 4.64TIQLQGSFTIDD478 pKa = 3.09KK479 pKa = 8.46LTNTQTGEE487 pKa = 4.16EE488 pKa = 4.26VTATSFDD495 pKa = 3.41SSEE498 pKa = 3.79PHH500 pKa = 6.37TDD502 pKa = 2.71SNYY505 pKa = 8.54ITQEE509 pKa = 3.3EE510 pKa = 4.64WDD512 pKa = 3.56QLEE515 pKa = 4.22QQNKK519 pKa = 8.28EE520 pKa = 4.17LIEE523 pKa = 4.64KK524 pKa = 10.22YY525 pKa = 9.73EE526 pKa = 3.97QSQSGGGLDD535 pKa = 4.4LGQFDD540 pKa = 4.01MFGIPGEE547 pKa = 4.08IVAVGVAALVGLGVLGNNN565 pKa = 4.05
MM1 pKa = 7.55GRR3 pKa = 11.84RR4 pKa = 11.84TFVKK8 pKa = 10.51GLGAAGAATVGLSVDD23 pKa = 4.34DD24 pKa = 5.68GFAQDD29 pKa = 3.91AQAIAPLVGVGLAAGAVGVGWALRR53 pKa = 11.84EE54 pKa = 3.93FEE56 pKa = 4.85IVGSDD61 pKa = 3.65APPEE65 pKa = 4.3GLTADD70 pKa = 3.62ALKK73 pKa = 10.31QQVYY77 pKa = 7.21QTAKK81 pKa = 8.92TRR83 pKa = 11.84KK84 pKa = 7.73STNASTIVDD93 pKa = 3.75NQNILDD99 pKa = 4.23GVKK102 pKa = 8.61HH103 pKa = 5.26TAYY106 pKa = 9.75TDD108 pKa = 3.32AKK110 pKa = 9.46IAAIEE115 pKa = 3.95EE116 pKa = 4.46LNAGSAEE123 pKa = 4.21SAVLDD128 pKa = 4.02AATTEE133 pKa = 4.3VNSYY137 pKa = 8.69LTTVQSNFLKK147 pKa = 9.59TWNEE151 pKa = 3.9SVAEE155 pKa = 4.06LDD157 pKa = 4.39SILSTVVNHH166 pKa = 6.72PDD168 pKa = 2.91IGKK171 pKa = 10.49GDD173 pKa = 3.92VFLMLNGSDD182 pKa = 3.54NTIEE186 pKa = 5.01DD187 pKa = 4.15LLANPSGSTDD197 pKa = 3.02ATSFTLADD205 pKa = 3.87GTTMSVGTVEE215 pKa = 4.02VDD217 pKa = 2.94RR218 pKa = 11.84GTEE221 pKa = 3.77SYY223 pKa = 11.09YY224 pKa = 10.34YY225 pKa = 10.72DD226 pKa = 3.98PMSGLVGDD234 pKa = 5.65LGDD237 pKa = 4.7LKK239 pKa = 11.3NGGPTVQYY247 pKa = 10.7DD248 pKa = 3.95GNSLVYY254 pKa = 10.65LNASNWKK261 pKa = 9.34PIYY264 pKa = 10.52DD265 pKa = 3.79EE266 pKa = 4.63MDD268 pKa = 3.1TVLQNVRR275 pKa = 11.84SGISTWVSNVYY286 pKa = 11.0GDD288 pKa = 3.81VQSGEE293 pKa = 4.29IEE295 pKa = 4.27VSDD298 pKa = 3.65LVTPRR303 pKa = 11.84EE304 pKa = 4.0RR305 pKa = 11.84AAMMAQEE312 pKa = 4.65EE313 pKa = 4.97GMSQAIADD321 pKa = 5.12LIALNVPVDD330 pKa = 3.84AEE332 pKa = 4.23RR333 pKa = 11.84EE334 pKa = 4.21ATITIQDD341 pKa = 3.56TGATLPGTFALTDD354 pKa = 3.64ASDD357 pKa = 4.16GPLEE361 pKa = 4.31SGKK364 pKa = 9.08TYY366 pKa = 10.79DD367 pKa = 3.71PSTFSGDD374 pKa = 3.59VYY376 pKa = 9.75FTADD380 pKa = 3.14MSLVEE385 pKa = 4.73GDD387 pKa = 2.89WTAYY391 pKa = 9.88QSGVDD396 pKa = 3.85GGNVTLTSEE405 pKa = 4.71PYY407 pKa = 10.31SGTAVEE413 pKa = 5.43LNTAANEE420 pKa = 4.25TVAVDD425 pKa = 3.73AGNWTATGNGTWYY438 pKa = 10.52HH439 pKa = 6.33DD440 pKa = 3.71VSPEE444 pKa = 4.05LEE446 pKa = 3.93TDD448 pKa = 2.85ITSIEE453 pKa = 4.21SARR456 pKa = 11.84FLSTAEE462 pKa = 3.68QTQYY466 pKa = 9.94EE467 pKa = 4.64TIQLQGSFTIDD478 pKa = 3.09KK479 pKa = 8.46LTNTQTGEE487 pKa = 4.16EE488 pKa = 4.26VTATSFDD495 pKa = 3.41SSEE498 pKa = 3.79PHH500 pKa = 6.37TDD502 pKa = 2.71SNYY505 pKa = 8.54ITQEE509 pKa = 3.3EE510 pKa = 4.64WDD512 pKa = 3.56QLEE515 pKa = 4.22QQNKK519 pKa = 8.28EE520 pKa = 4.17LIEE523 pKa = 4.64KK524 pKa = 10.22YY525 pKa = 9.73EE526 pKa = 3.97QSQSGGGLDD535 pKa = 4.4LGQFDD540 pKa = 4.01MFGIPGEE547 pKa = 4.08IVAVGVAALVGLGVLGNNN565 pKa = 4.05
Molecular weight: 59.61 kDa
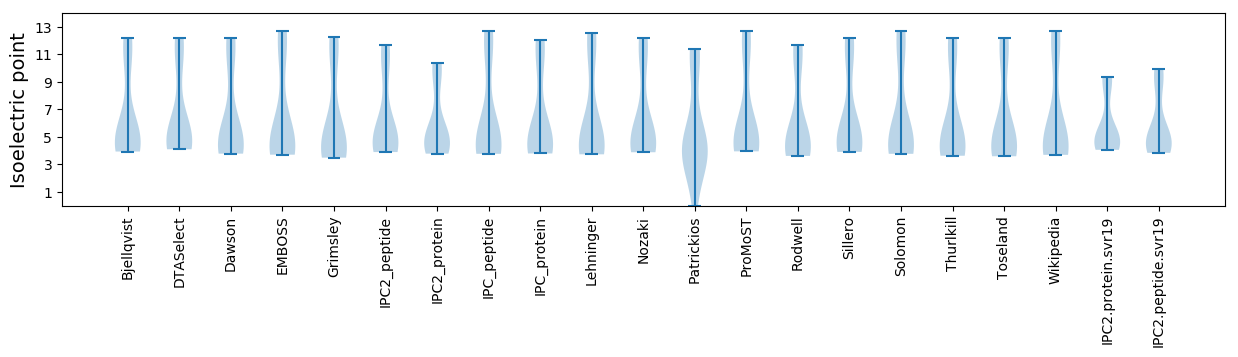
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9ABL7|H9ABL7_9VIRU ORF3 OS=Halorubrum pleomorphic virus 2 OX=1156719 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.63EE2 pKa = 4.65EE3 pKa = 3.72RR4 pKa = 11.84AKK6 pKa = 10.36QHH8 pKa = 5.94EE9 pKa = 4.74SPAARR14 pKa = 11.84AASYY18 pKa = 9.93PEE20 pKa = 3.92RR21 pKa = 11.84RR22 pKa = 11.84AKK24 pKa = 10.24RR25 pKa = 11.84AEE27 pKa = 4.12GYY29 pKa = 10.1RR30 pKa = 11.84DD31 pKa = 3.19GSRR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84QPGPASWSEE46 pKa = 3.56RR47 pKa = 11.84RR48 pKa = 11.84EE49 pKa = 3.95ISEE52 pKa = 3.84HH53 pKa = 4.24QQRR56 pKa = 11.84EE57 pKa = 4.08LRR59 pKa = 11.84GEE61 pKa = 3.78EE62 pKa = 4.01TRR64 pKa = 11.84RR65 pKa = 11.84QDD67 pKa = 3.59EE68 pKa = 4.5TRR70 pKa = 11.84MSTSSTRR77 pKa = 11.84LL78 pKa = 3.2
MM1 pKa = 7.63EE2 pKa = 4.65EE3 pKa = 3.72RR4 pKa = 11.84AKK6 pKa = 10.36QHH8 pKa = 5.94EE9 pKa = 4.74SPAARR14 pKa = 11.84AASYY18 pKa = 9.93PEE20 pKa = 3.92RR21 pKa = 11.84RR22 pKa = 11.84AKK24 pKa = 10.24RR25 pKa = 11.84AEE27 pKa = 4.12GYY29 pKa = 10.1RR30 pKa = 11.84DD31 pKa = 3.19GSRR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84QPGPASWSEE46 pKa = 3.56RR47 pKa = 11.84RR48 pKa = 11.84EE49 pKa = 3.95ISEE52 pKa = 3.84HH53 pKa = 4.24QQRR56 pKa = 11.84EE57 pKa = 4.08LRR59 pKa = 11.84GEE61 pKa = 3.78EE62 pKa = 4.01TRR64 pKa = 11.84RR65 pKa = 11.84QDD67 pKa = 3.59EE68 pKa = 4.5TRR70 pKa = 11.84MSTSSTRR77 pKa = 11.84LL78 pKa = 3.2
Molecular weight: 9.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3091 |
65 |
655 |
206.1 |
22.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.511 ± 0.6 | 1.003 ± 0.319 |
8.541 ± 1.147 | 6.632 ± 0.688 |
3.009 ± 0.356 | 8.25 ± 0.572 |
1.715 ± 0.382 | 3.365 ± 0.346 |
2.782 ± 0.479 | 9.059 ± 0.788 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.844 ± 0.212 | 2.653 ± 0.459 |
4.206 ± 0.377 | 3.235 ± 0.34 |
7.603 ± 1.216 | 7.667 ± 0.69 |
6.923 ± 0.723 | 7.732 ± 0.746 |
1.779 ± 0.183 | 2.491 ± 0.442 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
