
bacterium HR20
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
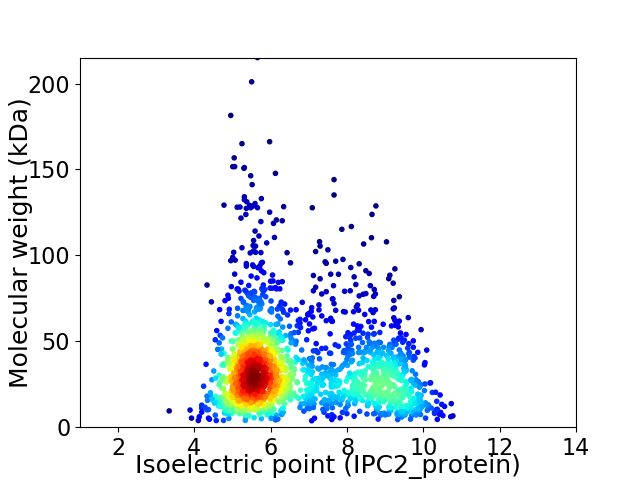
Virtual 2D-PAGE plot for 1779 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5XX39|A0A2H5XX39_9BACT Uncharacterized protein OS=bacterium HR20 OX=2035415 GN=HRbin20_01359 PE=4 SV=1
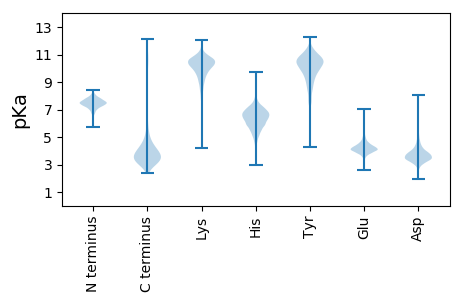
MM1 pKa = 7.97DD2 pKa = 4.68NKK4 pKa = 10.82DD5 pKa = 3.49VLTEE9 pKa = 3.89EE10 pKa = 5.11EE11 pKa = 4.08IAEE14 pKa = 4.72GYY16 pKa = 10.31ILTCQAHH23 pKa = 5.48PTAHH27 pKa = 6.14GVSVDD32 pKa = 3.42YY33 pKa = 11.1DD34 pKa = 3.35QQ35 pKa = 5.85
MM1 pKa = 7.97DD2 pKa = 4.68NKK4 pKa = 10.82DD5 pKa = 3.49VLTEE9 pKa = 3.89EE10 pKa = 5.11EE11 pKa = 4.08IAEE14 pKa = 4.72GYY16 pKa = 10.31ILTCQAHH23 pKa = 5.48PTAHH27 pKa = 6.14GVSVDD32 pKa = 3.42YY33 pKa = 11.1DD34 pKa = 3.35QQ35 pKa = 5.85
Molecular weight: 3.89 kDa
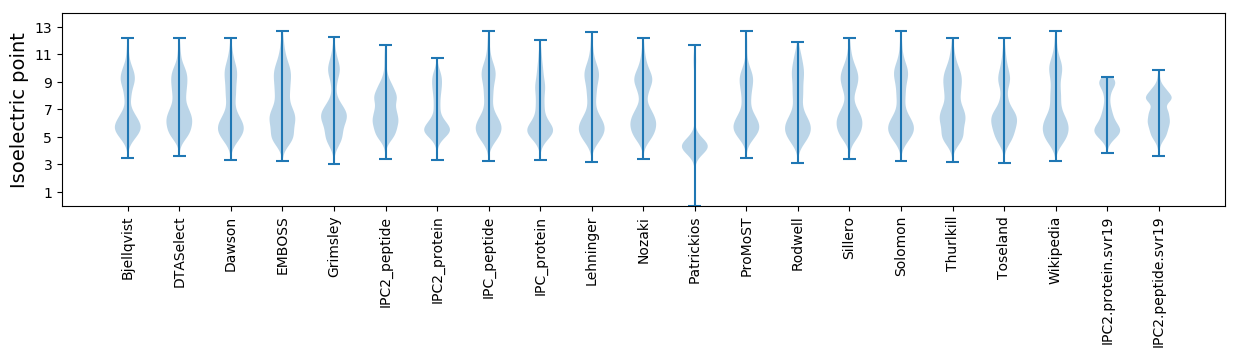
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5XUQ2|A0A2H5XUQ2_9BACT Uncharacterized protein OS=bacterium HR20 OX=2035415 GN=HRbin20_00484 PE=4 SV=1
MM1 pKa = 7.39INAASTEE8 pKa = 4.26PFCLEE13 pKa = 3.65ATTGGRR19 pKa = 11.84FLAGILPAAEE29 pKa = 3.98RR30 pKa = 11.84SVVSLPSQQKK40 pKa = 9.21RR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 6.71QTVRR47 pKa = 11.84KK48 pKa = 7.05TRR50 pKa = 11.84KK51 pKa = 4.83TTTVRR56 pKa = 11.84AKK58 pKa = 10.17RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84FAKK64 pKa = 10.09KK65 pKa = 8.16RR66 pKa = 11.84TAPNVSVRR74 pKa = 11.84PGLIRR79 pKa = 11.84GMTIVYY85 pKa = 7.7DD86 pKa = 3.82TLLVPGVIYY95 pKa = 10.54RR96 pKa = 11.84RR97 pKa = 11.84VDD99 pKa = 3.19VEE101 pKa = 4.39LADD104 pKa = 3.68STHH107 pKa = 6.51ALVHH111 pKa = 6.26AVTCVVRR118 pKa = 11.84NPRR121 pKa = 11.84YY122 pKa = 10.12GIEE125 pKa = 4.04LVQAHH130 pKa = 6.8DD131 pKa = 3.3RR132 pKa = 11.84LGRR135 pKa = 11.84LEE137 pKa = 4.25TLPALTRR144 pKa = 11.84RR145 pKa = 11.84LDD147 pKa = 3.47SAQGKK152 pKa = 9.18IVLAAINGSFWRR164 pKa = 11.84AGTRR168 pKa = 11.84QPLGISLSDD177 pKa = 3.68GEE179 pKa = 4.5IVAVTGAPWYY189 pKa = 10.33ALWLDD194 pKa = 3.84RR195 pKa = 11.84RR196 pKa = 11.84WRR198 pKa = 11.84PWLDD202 pKa = 2.97TGGVVATVHH211 pKa = 6.41LSDD214 pKa = 3.49GSTYY218 pKa = 10.53RR219 pKa = 11.84IDD221 pKa = 5.26AINASADD228 pKa = 3.2GVQLFNRR235 pKa = 11.84YY236 pKa = 9.49AGDD239 pKa = 4.31SIPVLPGQIPAEE251 pKa = 4.03LSTAQRR257 pKa = 11.84LITSDD262 pKa = 3.34TLDD265 pKa = 4.03PQWTSDD271 pKa = 3.42SLAAMLSTLQHH282 pKa = 4.44QWQRR286 pKa = 11.84DD287 pKa = 3.64LQKK290 pKa = 10.8PKK292 pKa = 10.9VLLRR296 pKa = 11.84YY297 pKa = 9.17LRR299 pKa = 11.84QPMVNQSTPCLVLRR313 pKa = 11.84TLDD316 pKa = 3.67SEE318 pKa = 4.34RR319 pKa = 11.84VEE321 pKa = 4.37VPLRR325 pKa = 11.84GCVLTVPRR333 pKa = 11.84GHH335 pKa = 7.12PLEE338 pKa = 4.28KK339 pKa = 10.32VIEE342 pKa = 4.27SGKK345 pKa = 10.25LKK347 pKa = 10.82RR348 pKa = 11.84GDD350 pKa = 3.66TVILQVRR357 pKa = 11.84TLRR360 pKa = 11.84NDD362 pKa = 3.96SIPFHH367 pKa = 5.7TAISGTPLLVKK378 pKa = 10.63NGTVKK383 pKa = 10.45PQLEE387 pKa = 4.18DD388 pKa = 3.13TLRR391 pKa = 11.84RR392 pKa = 11.84GSPFVEE398 pKa = 3.9QRR400 pKa = 11.84LSRR403 pKa = 11.84TAIGTDD409 pKa = 3.85VIQSVLYY416 pKa = 10.43LVTVEE421 pKa = 3.94QSPGRR426 pKa = 11.84SVGMSLRR433 pKa = 11.84EE434 pKa = 3.53LAQFMKK440 pKa = 10.64TFGASDD446 pKa = 3.78ALNLDD451 pKa = 4.11GGGSATMVVGDD462 pKa = 4.05RR463 pKa = 11.84CVVPPSGPSQCRR475 pKa = 11.84AVANALVIWRR485 pKa = 11.84RR486 pKa = 11.84KK487 pKa = 8.03PLSRR491 pKa = 5.17
MM1 pKa = 7.39INAASTEE8 pKa = 4.26PFCLEE13 pKa = 3.65ATTGGRR19 pKa = 11.84FLAGILPAAEE29 pKa = 3.98RR30 pKa = 11.84SVVSLPSQQKK40 pKa = 9.21RR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 6.71QTVRR47 pKa = 11.84KK48 pKa = 7.05TRR50 pKa = 11.84KK51 pKa = 4.83TTTVRR56 pKa = 11.84AKK58 pKa = 10.17RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84FAKK64 pKa = 10.09KK65 pKa = 8.16RR66 pKa = 11.84TAPNVSVRR74 pKa = 11.84PGLIRR79 pKa = 11.84GMTIVYY85 pKa = 7.7DD86 pKa = 3.82TLLVPGVIYY95 pKa = 10.54RR96 pKa = 11.84RR97 pKa = 11.84VDD99 pKa = 3.19VEE101 pKa = 4.39LADD104 pKa = 3.68STHH107 pKa = 6.51ALVHH111 pKa = 6.26AVTCVVRR118 pKa = 11.84NPRR121 pKa = 11.84YY122 pKa = 10.12GIEE125 pKa = 4.04LVQAHH130 pKa = 6.8DD131 pKa = 3.3RR132 pKa = 11.84LGRR135 pKa = 11.84LEE137 pKa = 4.25TLPALTRR144 pKa = 11.84RR145 pKa = 11.84LDD147 pKa = 3.47SAQGKK152 pKa = 9.18IVLAAINGSFWRR164 pKa = 11.84AGTRR168 pKa = 11.84QPLGISLSDD177 pKa = 3.68GEE179 pKa = 4.5IVAVTGAPWYY189 pKa = 10.33ALWLDD194 pKa = 3.84RR195 pKa = 11.84RR196 pKa = 11.84WRR198 pKa = 11.84PWLDD202 pKa = 2.97TGGVVATVHH211 pKa = 6.41LSDD214 pKa = 3.49GSTYY218 pKa = 10.53RR219 pKa = 11.84IDD221 pKa = 5.26AINASADD228 pKa = 3.2GVQLFNRR235 pKa = 11.84YY236 pKa = 9.49AGDD239 pKa = 4.31SIPVLPGQIPAEE251 pKa = 4.03LSTAQRR257 pKa = 11.84LITSDD262 pKa = 3.34TLDD265 pKa = 4.03PQWTSDD271 pKa = 3.42SLAAMLSTLQHH282 pKa = 4.44QWQRR286 pKa = 11.84DD287 pKa = 3.64LQKK290 pKa = 10.8PKK292 pKa = 10.9VLLRR296 pKa = 11.84YY297 pKa = 9.17LRR299 pKa = 11.84QPMVNQSTPCLVLRR313 pKa = 11.84TLDD316 pKa = 3.67SEE318 pKa = 4.34RR319 pKa = 11.84VEE321 pKa = 4.37VPLRR325 pKa = 11.84GCVLTVPRR333 pKa = 11.84GHH335 pKa = 7.12PLEE338 pKa = 4.28KK339 pKa = 10.32VIEE342 pKa = 4.27SGKK345 pKa = 10.25LKK347 pKa = 10.82RR348 pKa = 11.84GDD350 pKa = 3.66TVILQVRR357 pKa = 11.84TLRR360 pKa = 11.84NDD362 pKa = 3.96SIPFHH367 pKa = 5.7TAISGTPLLVKK378 pKa = 10.63NGTVKK383 pKa = 10.45PQLEE387 pKa = 4.18DD388 pKa = 3.13TLRR391 pKa = 11.84RR392 pKa = 11.84GSPFVEE398 pKa = 3.9QRR400 pKa = 11.84LSRR403 pKa = 11.84TAIGTDD409 pKa = 3.85VIQSVLYY416 pKa = 10.43LVTVEE421 pKa = 3.94QSPGRR426 pKa = 11.84SVGMSLRR433 pKa = 11.84EE434 pKa = 3.53LAQFMKK440 pKa = 10.64TFGASDD446 pKa = 3.78ALNLDD451 pKa = 4.11GGGSATMVVGDD462 pKa = 4.05RR463 pKa = 11.84CVVPPSGPSQCRR475 pKa = 11.84AVANALVIWRR485 pKa = 11.84RR486 pKa = 11.84KK487 pKa = 8.03PLSRR491 pKa = 5.17
Molecular weight: 53.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
604333 |
35 |
2059 |
339.7 |
37.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.458 ± 0.067 | 1.156 ± 0.025 |
4.871 ± 0.043 | 6.053 ± 0.07 |
3.594 ± 0.035 | 7.221 ± 0.056 |
2.181 ± 0.032 | 5.772 ± 0.041 |
2.405 ± 0.045 | 10.469 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.904 ± 0.024 | 2.65 ± 0.04 |
5.21 ± 0.037 | 4.033 ± 0.045 |
8.0 ± 0.054 | 6.002 ± 0.052 |
5.877 ± 0.062 | 7.671 ± 0.056 |
1.432 ± 0.025 | 3.043 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
