
Pothos latent virus (isolate Pigeonpea/India) (PoLV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Aureusvirus; Pothos latent virus
Average proteome isoelectric point is 8.23
Get precalculated fractions of proteins
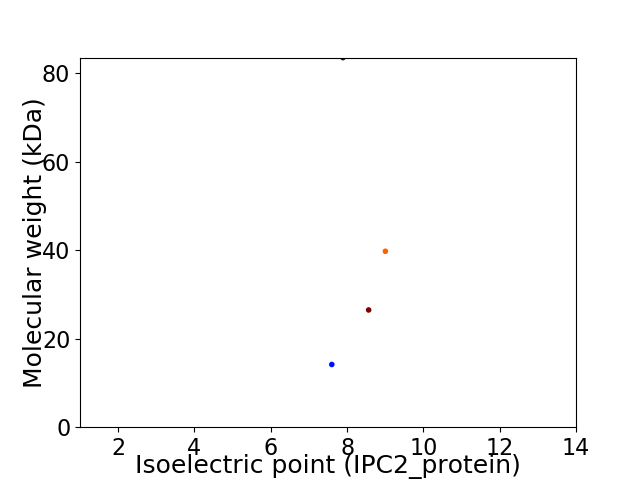
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9QBU1|P14_POLVP RNA silencing suppressor p14 OS=Pothos latent virus (isolate Pigeonpea/India) OX=652109 GN=ORF4 PE=4 SV=1
MM1 pKa = 7.57ANWCMSVLMFSFVTVYY17 pKa = 10.34TIVTSLGQGIRR28 pKa = 11.84RR29 pKa = 11.84GFRR32 pKa = 11.84AIVDD36 pKa = 3.86EE37 pKa = 4.35VEE39 pKa = 4.32NIYY42 pKa = 11.39GDD44 pKa = 3.35MCAGFKK50 pKa = 10.41VALSKK55 pKa = 11.2LSVSWIVVVGMIFVLNSLVVGLVPTLTMVAIIAACVGAKK94 pKa = 10.1YY95 pKa = 10.28GGRR98 pKa = 11.84VPRR101 pKa = 11.84YY102 pKa = 8.99IRR104 pKa = 11.84AHH106 pKa = 5.44VDD108 pKa = 4.04RR109 pKa = 11.84IRR111 pKa = 11.84KK112 pKa = 8.57SWEE115 pKa = 3.68KK116 pKa = 10.95GVDD119 pKa = 4.39DD120 pKa = 5.8DD121 pKa = 5.63DD122 pKa = 4.72CCAPLPEE129 pKa = 4.5TSLEE133 pKa = 4.05RR134 pKa = 11.84VPTKK138 pKa = 10.41LRR140 pKa = 11.84PRR142 pKa = 11.84LACKK146 pKa = 9.74IAVRR150 pKa = 11.84AIAKK154 pKa = 9.99VGLLKK159 pKa = 10.63RR160 pKa = 11.84SEE162 pKa = 4.34ANSMVYY168 pKa = 9.97QRR170 pKa = 11.84VCLDD174 pKa = 3.14VMEE177 pKa = 4.42SMKK180 pKa = 10.07MRR182 pKa = 11.84WHH184 pKa = 6.99DD185 pKa = 3.63RR186 pKa = 11.84LVILPQAVLACLEE199 pKa = 4.01RR200 pKa = 11.84PEE202 pKa = 4.2EE203 pKa = 4.11VEE205 pKa = 4.1EE206 pKa = 4.07VMKK209 pKa = 10.92AIEE212 pKa = 4.32ASCKK216 pKa = 10.41GRR218 pKa = 11.84FDD220 pKa = 3.98VYY222 pKa = 10.07XGCLLQHH229 pKa = 6.83RR230 pKa = 11.84GYY232 pKa = 8.77NTAIISAIPDD242 pKa = 3.55GVLLNHH248 pKa = 6.85EE249 pKa = 4.32GMVVRR254 pKa = 11.84KK255 pKa = 8.52GAPITTEE262 pKa = 3.85RR263 pKa = 11.84KK264 pKa = 7.59WYY266 pKa = 10.57SFAGYY271 pKa = 10.4ASTYY275 pKa = 10.04EE276 pKa = 4.24YY277 pKa = 10.18IVHH280 pKa = 6.48NSSLVNVCRR289 pKa = 11.84GLVEE293 pKa = 4.85RR294 pKa = 11.84VFCVVRR300 pKa = 11.84DD301 pKa = 4.08GKK303 pKa = 9.68LQRR306 pKa = 11.84PLRR309 pKa = 11.84PKK311 pKa = 10.72NNVFEE316 pKa = 4.67RR317 pKa = 11.84KK318 pKa = 9.26LGNVGRR324 pKa = 11.84RR325 pKa = 11.84ISSIVGYY332 pKa = 9.29CPAMTRR338 pKa = 11.84SEE340 pKa = 4.15FCEE343 pKa = 4.4SYY345 pKa = 10.65SGTRR349 pKa = 11.84RR350 pKa = 11.84ATYY353 pKa = 9.75EE354 pKa = 3.85RR355 pKa = 11.84ARR357 pKa = 11.84LSLDD361 pKa = 3.18VLPCTRR367 pKa = 11.84KK368 pKa = 9.84DD369 pKa = 3.24AYY371 pKa = 11.22LKK373 pKa = 10.07TFVKK377 pKa = 10.54AEE379 pKa = 4.43KK380 pKa = 10.4INITLKK386 pKa = 10.25PDD388 pKa = 3.82PAPRR392 pKa = 11.84VIQPRR397 pKa = 11.84DD398 pKa = 3.05PRR400 pKa = 11.84YY401 pKa = 8.24NVEE404 pKa = 3.6VGRR407 pKa = 11.84YY408 pKa = 7.77LKK410 pKa = 10.36PLEE413 pKa = 4.31PRR415 pKa = 11.84LMKK418 pKa = 10.74AIDD421 pKa = 3.85KK422 pKa = 10.71LWGEE426 pKa = 3.96KK427 pKa = 8.64TAIKK431 pKa = 10.27GYY433 pKa = 6.52TVEE436 pKa = 4.31KK437 pKa = 10.33VGEE440 pKa = 4.02ILHH443 pKa = 5.5QKK445 pKa = 8.49SLRR448 pKa = 11.84FKK450 pKa = 9.78NPCFVGLDD458 pKa = 3.29ASRR461 pKa = 11.84FDD463 pKa = 3.64QHH465 pKa = 7.34CSRR468 pKa = 11.84QALEE472 pKa = 4.51WEE474 pKa = 4.17HH475 pKa = 7.25SVYY478 pKa = 10.83NAIFRR483 pKa = 11.84DD484 pKa = 4.23PYY486 pKa = 10.98LSEE489 pKa = 4.3LLTWQIDD496 pKa = 4.22NVGTAYY502 pKa = 10.93LKK504 pKa = 10.95DD505 pKa = 3.2GFVRR509 pKa = 11.84YY510 pKa = 9.28RR511 pKa = 11.84VDD513 pKa = 3.25GCRR516 pKa = 11.84MSGDD520 pKa = 3.85MNTSMGNYY528 pKa = 10.0LIMSCLVYY536 pKa = 9.28QFCKK540 pKa = 10.63DD541 pKa = 3.15IGIDD545 pKa = 4.0ASLANCGDD553 pKa = 3.62DD554 pKa = 3.56CVLYY558 pKa = 10.76LEE560 pKa = 5.81KK561 pKa = 10.79EE562 pKa = 4.2DD563 pKa = 4.5LPKK566 pKa = 10.98LKK568 pKa = 10.46ALPDD572 pKa = 3.37WFGKK576 pKa = 9.1MGYY579 pKa = 8.44SMKK582 pKa = 10.2VEE584 pKa = 3.95KK585 pKa = 10.24PVFTVEE591 pKa = 4.44EE592 pKa = 4.2IEE594 pKa = 4.63FCQQHH599 pKa = 6.18PVQLSRR605 pKa = 11.84GYY607 pKa = 11.23VMVRR611 pKa = 11.84RR612 pKa = 11.84PDD614 pKa = 3.14VCLTKK619 pKa = 10.46DD620 pKa = 3.3LCVVRR625 pKa = 11.84GGMTTEE631 pKa = 4.28RR632 pKa = 11.84LQRR635 pKa = 11.84WLYY638 pKa = 8.84AQHH641 pKa = 7.37DD642 pKa = 4.28GGLALTGDD650 pKa = 4.49CPVLGAFYY658 pKa = 10.71RR659 pKa = 11.84RR660 pKa = 11.84FPSGEE665 pKa = 3.88SHH667 pKa = 6.95GEE669 pKa = 3.5LSEE672 pKa = 4.15YY673 pKa = 11.13SDD675 pKa = 3.14AHH677 pKa = 6.14KK678 pKa = 10.65FKK680 pKa = 10.88AGKK683 pKa = 9.85QYY685 pKa = 11.72GEE687 pKa = 4.15INSLARR693 pKa = 11.84YY694 pKa = 8.5SFWLAFGITPDD705 pKa = 3.32EE706 pKa = 3.92QLAIEE711 pKa = 4.68RR712 pKa = 11.84DD713 pKa = 3.68LMNWTPNVVPGSCDD727 pKa = 3.37PRR729 pKa = 11.84PTLLDD734 pKa = 3.46YY735 pKa = 11.46CSDD738 pKa = 3.48NN739 pKa = 3.65
MM1 pKa = 7.57ANWCMSVLMFSFVTVYY17 pKa = 10.34TIVTSLGQGIRR28 pKa = 11.84RR29 pKa = 11.84GFRR32 pKa = 11.84AIVDD36 pKa = 3.86EE37 pKa = 4.35VEE39 pKa = 4.32NIYY42 pKa = 11.39GDD44 pKa = 3.35MCAGFKK50 pKa = 10.41VALSKK55 pKa = 11.2LSVSWIVVVGMIFVLNSLVVGLVPTLTMVAIIAACVGAKK94 pKa = 10.1YY95 pKa = 10.28GGRR98 pKa = 11.84VPRR101 pKa = 11.84YY102 pKa = 8.99IRR104 pKa = 11.84AHH106 pKa = 5.44VDD108 pKa = 4.04RR109 pKa = 11.84IRR111 pKa = 11.84KK112 pKa = 8.57SWEE115 pKa = 3.68KK116 pKa = 10.95GVDD119 pKa = 4.39DD120 pKa = 5.8DD121 pKa = 5.63DD122 pKa = 4.72CCAPLPEE129 pKa = 4.5TSLEE133 pKa = 4.05RR134 pKa = 11.84VPTKK138 pKa = 10.41LRR140 pKa = 11.84PRR142 pKa = 11.84LACKK146 pKa = 9.74IAVRR150 pKa = 11.84AIAKK154 pKa = 9.99VGLLKK159 pKa = 10.63RR160 pKa = 11.84SEE162 pKa = 4.34ANSMVYY168 pKa = 9.97QRR170 pKa = 11.84VCLDD174 pKa = 3.14VMEE177 pKa = 4.42SMKK180 pKa = 10.07MRR182 pKa = 11.84WHH184 pKa = 6.99DD185 pKa = 3.63RR186 pKa = 11.84LVILPQAVLACLEE199 pKa = 4.01RR200 pKa = 11.84PEE202 pKa = 4.2EE203 pKa = 4.11VEE205 pKa = 4.1EE206 pKa = 4.07VMKK209 pKa = 10.92AIEE212 pKa = 4.32ASCKK216 pKa = 10.41GRR218 pKa = 11.84FDD220 pKa = 3.98VYY222 pKa = 10.07XGCLLQHH229 pKa = 6.83RR230 pKa = 11.84GYY232 pKa = 8.77NTAIISAIPDD242 pKa = 3.55GVLLNHH248 pKa = 6.85EE249 pKa = 4.32GMVVRR254 pKa = 11.84KK255 pKa = 8.52GAPITTEE262 pKa = 3.85RR263 pKa = 11.84KK264 pKa = 7.59WYY266 pKa = 10.57SFAGYY271 pKa = 10.4ASTYY275 pKa = 10.04EE276 pKa = 4.24YY277 pKa = 10.18IVHH280 pKa = 6.48NSSLVNVCRR289 pKa = 11.84GLVEE293 pKa = 4.85RR294 pKa = 11.84VFCVVRR300 pKa = 11.84DD301 pKa = 4.08GKK303 pKa = 9.68LQRR306 pKa = 11.84PLRR309 pKa = 11.84PKK311 pKa = 10.72NNVFEE316 pKa = 4.67RR317 pKa = 11.84KK318 pKa = 9.26LGNVGRR324 pKa = 11.84RR325 pKa = 11.84ISSIVGYY332 pKa = 9.29CPAMTRR338 pKa = 11.84SEE340 pKa = 4.15FCEE343 pKa = 4.4SYY345 pKa = 10.65SGTRR349 pKa = 11.84RR350 pKa = 11.84ATYY353 pKa = 9.75EE354 pKa = 3.85RR355 pKa = 11.84ARR357 pKa = 11.84LSLDD361 pKa = 3.18VLPCTRR367 pKa = 11.84KK368 pKa = 9.84DD369 pKa = 3.24AYY371 pKa = 11.22LKK373 pKa = 10.07TFVKK377 pKa = 10.54AEE379 pKa = 4.43KK380 pKa = 10.4INITLKK386 pKa = 10.25PDD388 pKa = 3.82PAPRR392 pKa = 11.84VIQPRR397 pKa = 11.84DD398 pKa = 3.05PRR400 pKa = 11.84YY401 pKa = 8.24NVEE404 pKa = 3.6VGRR407 pKa = 11.84YY408 pKa = 7.77LKK410 pKa = 10.36PLEE413 pKa = 4.31PRR415 pKa = 11.84LMKK418 pKa = 10.74AIDD421 pKa = 3.85KK422 pKa = 10.71LWGEE426 pKa = 3.96KK427 pKa = 8.64TAIKK431 pKa = 10.27GYY433 pKa = 6.52TVEE436 pKa = 4.31KK437 pKa = 10.33VGEE440 pKa = 4.02ILHH443 pKa = 5.5QKK445 pKa = 8.49SLRR448 pKa = 11.84FKK450 pKa = 9.78NPCFVGLDD458 pKa = 3.29ASRR461 pKa = 11.84FDD463 pKa = 3.64QHH465 pKa = 7.34CSRR468 pKa = 11.84QALEE472 pKa = 4.51WEE474 pKa = 4.17HH475 pKa = 7.25SVYY478 pKa = 10.83NAIFRR483 pKa = 11.84DD484 pKa = 4.23PYY486 pKa = 10.98LSEE489 pKa = 4.3LLTWQIDD496 pKa = 4.22NVGTAYY502 pKa = 10.93LKK504 pKa = 10.95DD505 pKa = 3.2GFVRR509 pKa = 11.84YY510 pKa = 9.28RR511 pKa = 11.84VDD513 pKa = 3.25GCRR516 pKa = 11.84MSGDD520 pKa = 3.85MNTSMGNYY528 pKa = 10.0LIMSCLVYY536 pKa = 9.28QFCKK540 pKa = 10.63DD541 pKa = 3.15IGIDD545 pKa = 4.0ASLANCGDD553 pKa = 3.62DD554 pKa = 3.56CVLYY558 pKa = 10.76LEE560 pKa = 5.81KK561 pKa = 10.79EE562 pKa = 4.2DD563 pKa = 4.5LPKK566 pKa = 10.98LKK568 pKa = 10.46ALPDD572 pKa = 3.37WFGKK576 pKa = 9.1MGYY579 pKa = 8.44SMKK582 pKa = 10.2VEE584 pKa = 3.95KK585 pKa = 10.24PVFTVEE591 pKa = 4.44EE592 pKa = 4.2IEE594 pKa = 4.63FCQQHH599 pKa = 6.18PVQLSRR605 pKa = 11.84GYY607 pKa = 11.23VMVRR611 pKa = 11.84RR612 pKa = 11.84PDD614 pKa = 3.14VCLTKK619 pKa = 10.46DD620 pKa = 3.3LCVVRR625 pKa = 11.84GGMTTEE631 pKa = 4.28RR632 pKa = 11.84LQRR635 pKa = 11.84WLYY638 pKa = 8.84AQHH641 pKa = 7.37DD642 pKa = 4.28GGLALTGDD650 pKa = 4.49CPVLGAFYY658 pKa = 10.71RR659 pKa = 11.84RR660 pKa = 11.84FPSGEE665 pKa = 3.88SHH667 pKa = 6.95GEE669 pKa = 3.5LSEE672 pKa = 4.15YY673 pKa = 11.13SDD675 pKa = 3.14AHH677 pKa = 6.14KK678 pKa = 10.65FKK680 pKa = 10.88AGKK683 pKa = 9.85QYY685 pKa = 11.72GEE687 pKa = 4.15INSLARR693 pKa = 11.84YY694 pKa = 8.5SFWLAFGITPDD705 pKa = 3.32EE706 pKa = 3.92QLAIEE711 pKa = 4.68RR712 pKa = 11.84DD713 pKa = 3.68LMNWTPNVVPGSCDD727 pKa = 3.37PRR729 pKa = 11.84PTLLDD734 pKa = 3.46YY735 pKa = 11.46CSDD738 pKa = 3.48NN739 pKa = 3.65
Molecular weight: 83.59 kDa
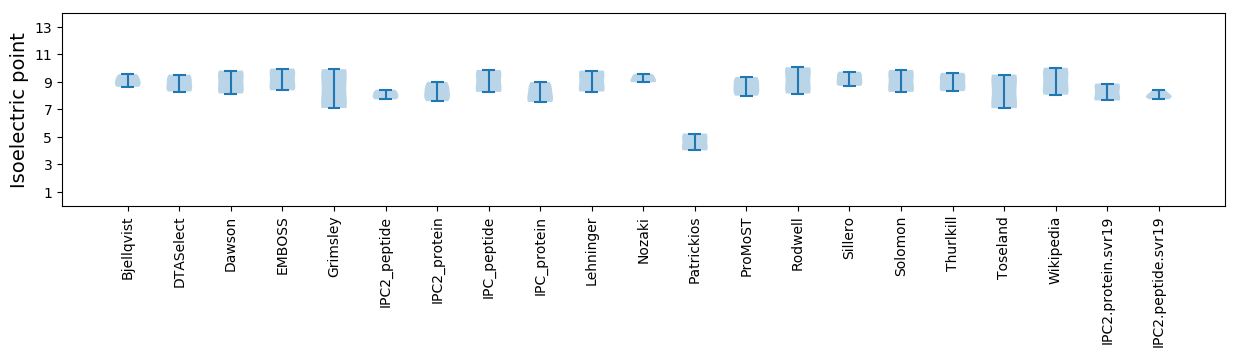
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9QBU3|CAPSD_POLVP Capsid protein OS=Pothos latent virus (isolate Pigeonpea/India) OX=652109 GN=ORF2 PE=3 SV=1
MM1 pKa = 7.59ALVKK5 pKa = 10.53RR6 pKa = 11.84NNNMALIATEE16 pKa = 4.13AGKK19 pKa = 9.96LAAVKK24 pKa = 10.09AGQVMLSPAGRR35 pKa = 11.84EE36 pKa = 4.14LIWNGVNWVRR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 10.2LGRR51 pKa = 11.84SKK53 pKa = 10.83KK54 pKa = 10.15SDD56 pKa = 3.67VILHH60 pKa = 6.32PGVLPGAIAAPVANTRR76 pKa = 11.84IIRR79 pKa = 11.84ASKK82 pKa = 10.4PKK84 pKa = 8.67FTRR87 pKa = 11.84SKK89 pKa = 10.13GSVTIAHH96 pKa = 6.98RR97 pKa = 11.84EE98 pKa = 3.74LLGQFNNSSGLVVNGGVSGNLYY120 pKa = 10.34RR121 pKa = 11.84INPSNPVVFPWLQGIAASFDD141 pKa = 3.23QYY143 pKa = 11.72KK144 pKa = 9.98FDD146 pKa = 4.33RR147 pKa = 11.84VQLQYY152 pKa = 11.56VPMCATTEE160 pKa = 4.0TGRR163 pKa = 11.84VAIYY167 pKa = 10.17FDD169 pKa = 4.79KK170 pKa = 10.84DD171 pKa = 3.53QQDD174 pKa = 3.61VEE176 pKa = 4.15PADD179 pKa = 3.91RR180 pKa = 11.84VEE182 pKa = 4.15LANMGHH188 pKa = 5.31WTEE191 pKa = 4.04SAPWCEE197 pKa = 4.01STLNVPVDD205 pKa = 3.46NVKK208 pKa = 10.82RR209 pKa = 11.84FMNDD213 pKa = 2.15NTTTDD218 pKa = 3.26RR219 pKa = 11.84KK220 pKa = 10.46LVDD223 pKa = 4.24LGQIGLATYY232 pKa = 10.59GGGSTNPVGDD242 pKa = 4.1LFIHH246 pKa = 4.8YY247 pKa = 7.38TITLFEE253 pKa = 4.42PQPLASLVEE262 pKa = 4.45TEE264 pKa = 4.05QSGAGAAPFGTNLVTVSSNATTTIITFRR292 pKa = 11.84GPGVYY297 pKa = 10.49LLALSQRR304 pKa = 11.84AASFTTLVTAGGAVVNSHH322 pKa = 5.28TTLFSGPAYY331 pKa = 10.33QSIANITATIPGGSITYY348 pKa = 10.42NGTLFGNYY356 pKa = 7.23TLQVTRR362 pKa = 11.84AKK364 pKa = 10.24ISNNATLII372 pKa = 3.97
MM1 pKa = 7.59ALVKK5 pKa = 10.53RR6 pKa = 11.84NNNMALIATEE16 pKa = 4.13AGKK19 pKa = 9.96LAAVKK24 pKa = 10.09AGQVMLSPAGRR35 pKa = 11.84EE36 pKa = 4.14LIWNGVNWVRR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 10.2LGRR51 pKa = 11.84SKK53 pKa = 10.83KK54 pKa = 10.15SDD56 pKa = 3.67VILHH60 pKa = 6.32PGVLPGAIAAPVANTRR76 pKa = 11.84IIRR79 pKa = 11.84ASKK82 pKa = 10.4PKK84 pKa = 8.67FTRR87 pKa = 11.84SKK89 pKa = 10.13GSVTIAHH96 pKa = 6.98RR97 pKa = 11.84EE98 pKa = 3.74LLGQFNNSSGLVVNGGVSGNLYY120 pKa = 10.34RR121 pKa = 11.84INPSNPVVFPWLQGIAASFDD141 pKa = 3.23QYY143 pKa = 11.72KK144 pKa = 9.98FDD146 pKa = 4.33RR147 pKa = 11.84VQLQYY152 pKa = 11.56VPMCATTEE160 pKa = 4.0TGRR163 pKa = 11.84VAIYY167 pKa = 10.17FDD169 pKa = 4.79KK170 pKa = 10.84DD171 pKa = 3.53QQDD174 pKa = 3.61VEE176 pKa = 4.15PADD179 pKa = 3.91RR180 pKa = 11.84VEE182 pKa = 4.15LANMGHH188 pKa = 5.31WTEE191 pKa = 4.04SAPWCEE197 pKa = 4.01STLNVPVDD205 pKa = 3.46NVKK208 pKa = 10.82RR209 pKa = 11.84FMNDD213 pKa = 2.15NTTTDD218 pKa = 3.26RR219 pKa = 11.84KK220 pKa = 10.46LVDD223 pKa = 4.24LGQIGLATYY232 pKa = 10.59GGGSTNPVGDD242 pKa = 4.1LFIHH246 pKa = 4.8YY247 pKa = 7.38TITLFEE253 pKa = 4.42PQPLASLVEE262 pKa = 4.45TEE264 pKa = 4.05QSGAGAAPFGTNLVTVSSNATTTIITFRR292 pKa = 11.84GPGVYY297 pKa = 10.49LLALSQRR304 pKa = 11.84AASFTTLVTAGGAVVNSHH322 pKa = 5.28TTLFSGPAYY331 pKa = 10.33QSIANITATIPGGSITYY348 pKa = 10.42NGTLFGNYY356 pKa = 7.23TLQVTRR362 pKa = 11.84AKK364 pKa = 10.24ISNNATLII372 pKa = 3.97
Molecular weight: 39.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1483 |
130 |
739 |
370.8 |
41.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.676 ± 1.122 | 2.495 ± 0.948 |
4.316 ± 0.657 | 4.99 ± 0.67 |
3.574 ± 0.166 | 8.159 ± 0.677 |
1.821 ± 0.335 | 4.922 ± 0.265 |
5.057 ± 0.675 | 8.699 ± 0.49 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.495 ± 0.412 | 3.978 ± 0.934 |
4.99 ± 0.306 | 3.709 ± 0.922 |
6.541 ± 0.885 | 7.822 ± 1.782 |
5.934 ± 1.315 | 8.833 ± 0.502 |
1.416 ± 0.142 | 3.506 ± 0.582 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
