
Acholeplasma phage L2 (Bacteriophage L2)
Taxonomy: Viruses; Plasmaviridae; Plasmavirus
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
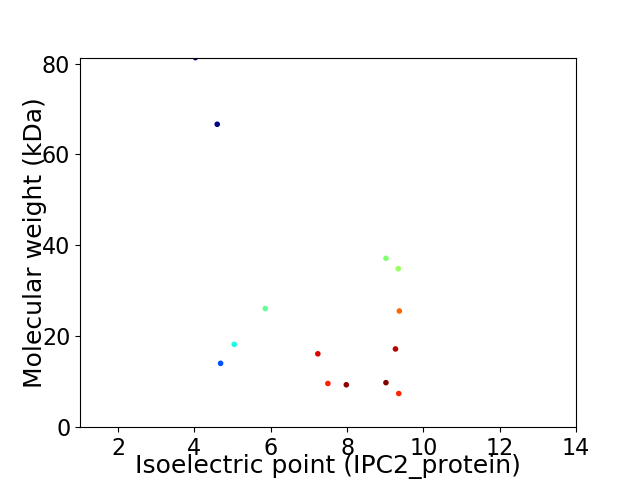
Virtual 2D-PAGE plot for 14 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P42549|YO14_BPL2 Uncharacterized 26.1 kDa protein OS=Acholeplasma phage L2 OX=46014 PE=3 SV=1
MM1 pKa = 7.52KK2 pKa = 10.52KK3 pKa = 10.46LLTSLMLSTASFMLLLTVSNKK24 pKa = 9.85AYY26 pKa = 10.11AYY28 pKa = 9.99EE29 pKa = 4.04EE30 pKa = 4.64TEE32 pKa = 4.04TFTFEE37 pKa = 4.02IYY39 pKa = 11.29NMNSVVANPGLYY51 pKa = 10.2NYY53 pKa = 10.64LVDD56 pKa = 3.83LTDD59 pKa = 4.25GYY61 pKa = 11.78GYY63 pKa = 11.25GQMQHH68 pKa = 4.74VTVDD72 pKa = 3.52ISLLDD77 pKa = 3.57YY78 pKa = 9.34NTAYY82 pKa = 10.86LEE84 pKa = 4.3KK85 pKa = 10.59NLNGFNLFFTSDD97 pKa = 2.91TGVYY101 pKa = 10.08EE102 pKa = 4.21FGIDD106 pKa = 3.13ASGNIEE112 pKa = 4.47LLNDD116 pKa = 3.9PVAHH120 pKa = 7.15DD121 pKa = 4.51EE122 pKa = 4.22YY123 pKa = 11.5SLTFTGLNNAYY134 pKa = 9.94QDD136 pKa = 3.61LFYY139 pKa = 11.42EE140 pKa = 4.5EE141 pKa = 4.48IVVEE145 pKa = 4.35FEE147 pKa = 3.97IVSFDD152 pKa = 3.42SLEE155 pKa = 4.37GVGTLDD161 pKa = 3.47PNPYY165 pKa = 9.86VVLSEE170 pKa = 4.7LLTEE174 pKa = 4.05NYY176 pKa = 9.54EE177 pKa = 4.22YY178 pKa = 11.37NEE180 pKa = 4.32TIAPFIWVNDD190 pKa = 3.53VQYY193 pKa = 10.95QNYY196 pKa = 9.12YY197 pKa = 9.48SVYY200 pKa = 10.59DD201 pKa = 3.58IGSDD205 pKa = 3.35LALIEE210 pKa = 4.51IDD212 pKa = 4.42GPTLNIYY219 pKa = 9.84SDD221 pKa = 3.36RR222 pKa = 11.84FEE224 pKa = 4.85EE225 pKa = 4.27YY226 pKa = 10.96NEE228 pKa = 4.12TFDD231 pKa = 4.22VGDD234 pKa = 3.6EE235 pKa = 3.71FRR237 pKa = 11.84IIFKK241 pKa = 10.11IYY243 pKa = 8.73KK244 pKa = 8.2TWTQAHH250 pKa = 7.17DD251 pKa = 3.56YY252 pKa = 11.23GRR254 pKa = 11.84IRR256 pKa = 11.84TNASTLYY263 pKa = 10.73LNQQSDD269 pKa = 3.63TSALLSHH276 pKa = 6.53SEE278 pKa = 3.98STNVINVNNIAVGDD292 pKa = 4.36EE293 pKa = 3.97IVMQSPYY300 pKa = 10.4TDD302 pKa = 4.27FSVSYY307 pKa = 10.31GNVGFPRR314 pKa = 11.84TYY316 pKa = 10.6LVEE319 pKa = 4.09QEE321 pKa = 4.45NIAAVKK327 pKa = 9.26MIKK330 pKa = 10.25NSSDD334 pKa = 2.97NTNYY338 pKa = 9.94TFILFYY344 pKa = 10.11WEE346 pKa = 4.03NGSVVSRR353 pKa = 11.84TVNNINLANRR363 pKa = 11.84NSLNIKK369 pKa = 9.92FDD371 pKa = 3.3GGFRR375 pKa = 11.84KK376 pKa = 9.9ALNYY380 pKa = 10.08SDD382 pKa = 5.0VMVANVDD389 pKa = 4.04LLTPLEE395 pKa = 4.54DD396 pKa = 4.17FLPNMVAWDD405 pKa = 5.07AIDD408 pKa = 5.04GDD410 pKa = 3.88ISDD413 pKa = 5.37SIVITDD419 pKa = 3.67NDD421 pKa = 4.62GYY423 pKa = 11.52SPDD426 pKa = 3.38TVGIYY431 pKa = 10.06NVEE434 pKa = 4.0FSVTNSNGQTSSIIAPVHH452 pKa = 5.51VVDD455 pKa = 5.36IVNPVINGVSDD466 pKa = 3.88TVHH469 pKa = 6.48ISYY472 pKa = 10.42DD473 pKa = 3.16QTFNVTNWVNSLTVSDD489 pKa = 4.31NYY491 pKa = 9.98YY492 pKa = 9.31TGLSISIKK500 pKa = 9.89EE501 pKa = 3.93NTYY504 pKa = 9.14TVNKK508 pKa = 9.88NKK510 pKa = 10.51LGTYY514 pKa = 9.64KK515 pKa = 9.4ITVQAVDD522 pKa = 3.43PSGNIGTLTRR532 pKa = 11.84TIVVNDD538 pKa = 4.93GIGPVFNGINTITASINEE556 pKa = 4.7NITVEE561 pKa = 4.1QIKK564 pKa = 10.75AGLAAIDD571 pKa = 5.08AIDD574 pKa = 4.18GNVTTSIVVDD584 pKa = 3.71SDD586 pKa = 3.64NLTGKK591 pKa = 10.51ANTVGVYY598 pKa = 9.58EE599 pKa = 4.36VVFRR603 pKa = 11.84AVDD606 pKa = 3.12AAGNQTFHH614 pKa = 6.44TVTVSIVASPPGFYY628 pKa = 10.06ILNSNSVRR636 pKa = 11.84LLPGANLTIEE646 pKa = 4.25QILNILNASDD656 pKa = 4.44AEE658 pKa = 4.48NISTNYY664 pKa = 8.05TVSVPGTYY672 pKa = 10.35NLSFTLYY679 pKa = 10.42GEE681 pKa = 4.21SHH683 pKa = 5.52QVSITVLGQNDD694 pKa = 4.76SIIPTPVIPGEE705 pKa = 4.28SNPGFNITYY714 pKa = 10.41ALIIGLSAIALLATTVTILNKK735 pKa = 9.61KK736 pKa = 9.61RR737 pKa = 11.84KK738 pKa = 9.02
MM1 pKa = 7.52KK2 pKa = 10.52KK3 pKa = 10.46LLTSLMLSTASFMLLLTVSNKK24 pKa = 9.85AYY26 pKa = 10.11AYY28 pKa = 9.99EE29 pKa = 4.04EE30 pKa = 4.64TEE32 pKa = 4.04TFTFEE37 pKa = 4.02IYY39 pKa = 11.29NMNSVVANPGLYY51 pKa = 10.2NYY53 pKa = 10.64LVDD56 pKa = 3.83LTDD59 pKa = 4.25GYY61 pKa = 11.78GYY63 pKa = 11.25GQMQHH68 pKa = 4.74VTVDD72 pKa = 3.52ISLLDD77 pKa = 3.57YY78 pKa = 9.34NTAYY82 pKa = 10.86LEE84 pKa = 4.3KK85 pKa = 10.59NLNGFNLFFTSDD97 pKa = 2.91TGVYY101 pKa = 10.08EE102 pKa = 4.21FGIDD106 pKa = 3.13ASGNIEE112 pKa = 4.47LLNDD116 pKa = 3.9PVAHH120 pKa = 7.15DD121 pKa = 4.51EE122 pKa = 4.22YY123 pKa = 11.5SLTFTGLNNAYY134 pKa = 9.94QDD136 pKa = 3.61LFYY139 pKa = 11.42EE140 pKa = 4.5EE141 pKa = 4.48IVVEE145 pKa = 4.35FEE147 pKa = 3.97IVSFDD152 pKa = 3.42SLEE155 pKa = 4.37GVGTLDD161 pKa = 3.47PNPYY165 pKa = 9.86VVLSEE170 pKa = 4.7LLTEE174 pKa = 4.05NYY176 pKa = 9.54EE177 pKa = 4.22YY178 pKa = 11.37NEE180 pKa = 4.32TIAPFIWVNDD190 pKa = 3.53VQYY193 pKa = 10.95QNYY196 pKa = 9.12YY197 pKa = 9.48SVYY200 pKa = 10.59DD201 pKa = 3.58IGSDD205 pKa = 3.35LALIEE210 pKa = 4.51IDD212 pKa = 4.42GPTLNIYY219 pKa = 9.84SDD221 pKa = 3.36RR222 pKa = 11.84FEE224 pKa = 4.85EE225 pKa = 4.27YY226 pKa = 10.96NEE228 pKa = 4.12TFDD231 pKa = 4.22VGDD234 pKa = 3.6EE235 pKa = 3.71FRR237 pKa = 11.84IIFKK241 pKa = 10.11IYY243 pKa = 8.73KK244 pKa = 8.2TWTQAHH250 pKa = 7.17DD251 pKa = 3.56YY252 pKa = 11.23GRR254 pKa = 11.84IRR256 pKa = 11.84TNASTLYY263 pKa = 10.73LNQQSDD269 pKa = 3.63TSALLSHH276 pKa = 6.53SEE278 pKa = 3.98STNVINVNNIAVGDD292 pKa = 4.36EE293 pKa = 3.97IVMQSPYY300 pKa = 10.4TDD302 pKa = 4.27FSVSYY307 pKa = 10.31GNVGFPRR314 pKa = 11.84TYY316 pKa = 10.6LVEE319 pKa = 4.09QEE321 pKa = 4.45NIAAVKK327 pKa = 9.26MIKK330 pKa = 10.25NSSDD334 pKa = 2.97NTNYY338 pKa = 9.94TFILFYY344 pKa = 10.11WEE346 pKa = 4.03NGSVVSRR353 pKa = 11.84TVNNINLANRR363 pKa = 11.84NSLNIKK369 pKa = 9.92FDD371 pKa = 3.3GGFRR375 pKa = 11.84KK376 pKa = 9.9ALNYY380 pKa = 10.08SDD382 pKa = 5.0VMVANVDD389 pKa = 4.04LLTPLEE395 pKa = 4.54DD396 pKa = 4.17FLPNMVAWDD405 pKa = 5.07AIDD408 pKa = 5.04GDD410 pKa = 3.88ISDD413 pKa = 5.37SIVITDD419 pKa = 3.67NDD421 pKa = 4.62GYY423 pKa = 11.52SPDD426 pKa = 3.38TVGIYY431 pKa = 10.06NVEE434 pKa = 4.0FSVTNSNGQTSSIIAPVHH452 pKa = 5.51VVDD455 pKa = 5.36IVNPVINGVSDD466 pKa = 3.88TVHH469 pKa = 6.48ISYY472 pKa = 10.42DD473 pKa = 3.16QTFNVTNWVNSLTVSDD489 pKa = 4.31NYY491 pKa = 9.98YY492 pKa = 9.31TGLSISIKK500 pKa = 9.89EE501 pKa = 3.93NTYY504 pKa = 9.14TVNKK508 pKa = 9.88NKK510 pKa = 10.51LGTYY514 pKa = 9.64KK515 pKa = 9.4ITVQAVDD522 pKa = 3.43PSGNIGTLTRR532 pKa = 11.84TIVVNDD538 pKa = 4.93GIGPVFNGINTITASINEE556 pKa = 4.7NITVEE561 pKa = 4.1QIKK564 pKa = 10.75AGLAAIDD571 pKa = 5.08AIDD574 pKa = 4.18GNVTTSIVVDD584 pKa = 3.71SDD586 pKa = 3.64NLTGKK591 pKa = 10.51ANTVGVYY598 pKa = 9.58EE599 pKa = 4.36VVFRR603 pKa = 11.84AVDD606 pKa = 3.12AAGNQTFHH614 pKa = 6.44TVTVSIVASPPGFYY628 pKa = 10.06ILNSNSVRR636 pKa = 11.84LLPGANLTIEE646 pKa = 4.25QILNILNASDD656 pKa = 4.44AEE658 pKa = 4.48NISTNYY664 pKa = 8.05TVSVPGTYY672 pKa = 10.35NLSFTLYY679 pKa = 10.42GEE681 pKa = 4.21SHH683 pKa = 5.52QVSITVLGQNDD694 pKa = 4.76SIIPTPVIPGEE705 pKa = 4.28SNPGFNITYY714 pKa = 10.41ALIIGLSAIALLATTVTILNKK735 pKa = 9.61KK736 pKa = 9.61RR737 pKa = 11.84KK738 pKa = 9.02
Molecular weight: 81.31 kDa
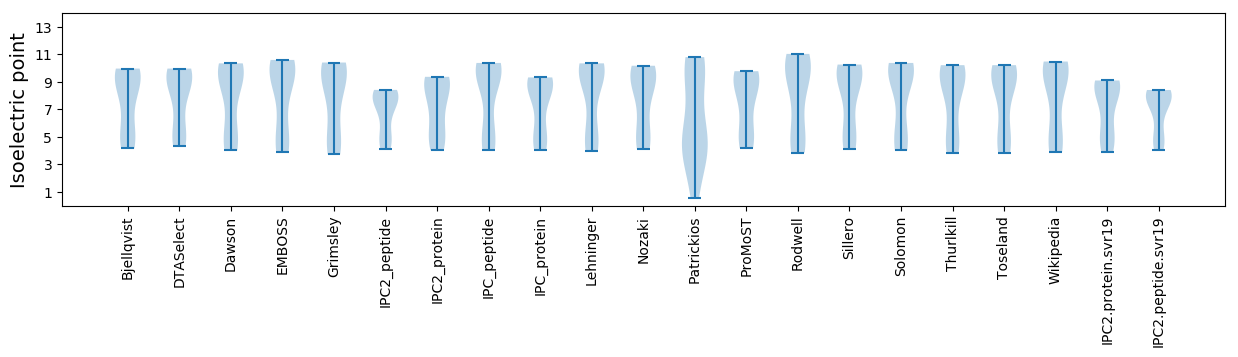
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P42541|YO06_BPL2 Uncharacterized 9.8 kDa protein OS=Acholeplasma phage L2 OX=46014 PE=4 SV=1
MM1 pKa = 7.38NIEE4 pKa = 4.18KK5 pKa = 10.76ALMNYY10 pKa = 9.87YY11 pKa = 10.37EE12 pKa = 4.53LTLASYY18 pKa = 10.82EE19 pKa = 4.21DD20 pKa = 3.58STNRR24 pKa = 11.84YY25 pKa = 8.16HH26 pKa = 7.19YY27 pKa = 10.36RR28 pKa = 11.84IVQHH32 pKa = 6.6LIKK35 pKa = 10.07GLKK38 pKa = 8.12YY39 pKa = 10.14IKK41 pKa = 10.09VKK43 pKa = 10.64KK44 pKa = 10.07LEE46 pKa = 4.33KK47 pKa = 10.06VDD49 pKa = 3.13ITIGYY54 pKa = 9.53KK55 pKa = 10.71LIDD58 pKa = 3.48YY59 pKa = 8.36LKK61 pKa = 10.2NHH63 pKa = 5.56THH65 pKa = 6.56NGNNSIKK72 pKa = 10.58KK73 pKa = 9.31IINYY77 pKa = 8.4LRR79 pKa = 11.84KK80 pKa = 9.74VMQHH84 pKa = 5.06YY85 pKa = 10.51RR86 pKa = 11.84ITTSIIDD93 pKa = 4.31LPHH96 pKa = 6.89LPNDD100 pKa = 3.77TKK102 pKa = 10.69PFEE105 pKa = 4.42RR106 pKa = 11.84FYY108 pKa = 11.19HH109 pKa = 7.2DD110 pKa = 4.35DD111 pKa = 3.86LEE113 pKa = 6.8LIMTYY118 pKa = 9.33TKK120 pKa = 10.97NLNSSKK126 pKa = 11.09NSITYY131 pKa = 10.16KK132 pKa = 10.61SFIRR136 pKa = 11.84LLLDD140 pKa = 3.25SGLRR144 pKa = 11.84VSEE147 pKa = 4.06ALNIKK152 pKa = 9.95ISDD155 pKa = 3.48MDD157 pKa = 4.08FKK159 pKa = 11.52NKK161 pKa = 9.59VIRR164 pKa = 11.84VLSSKK169 pKa = 7.56TRR171 pKa = 11.84KK172 pKa = 8.58QRR174 pKa = 11.84YY175 pKa = 8.83APFSSFSLKK184 pKa = 10.45YY185 pKa = 9.1IKK187 pKa = 10.21EE188 pKa = 4.28LIEE191 pKa = 3.98VNPKK195 pKa = 9.85RR196 pKa = 11.84DD197 pKa = 3.4YY198 pKa = 11.49LFYY201 pKa = 11.42NFIKK205 pKa = 10.58DD206 pKa = 3.33RR207 pKa = 11.84QTNKK211 pKa = 10.15NDD213 pKa = 2.59IKK215 pKa = 11.03LFYY218 pKa = 10.41KK219 pKa = 10.46RR220 pKa = 11.84LKK222 pKa = 10.12KK223 pKa = 9.99HH224 pKa = 6.2LNLEE228 pKa = 4.78RR229 pKa = 11.84IHH231 pKa = 5.08THH233 pKa = 5.76RR234 pKa = 11.84FRR236 pKa = 11.84KK237 pKa = 7.82TFASILIEE245 pKa = 4.23NGLNIDD251 pKa = 4.3DD252 pKa = 4.07LQKK255 pKa = 10.67IFDD258 pKa = 3.77HH259 pKa = 6.61SRR261 pKa = 11.84IEE263 pKa = 4.01TTIKK267 pKa = 10.3YY268 pKa = 8.37VQHH271 pKa = 5.74NEE273 pKa = 3.27KK274 pKa = 10.4RR275 pKa = 11.84ALQEE279 pKa = 3.82YY280 pKa = 10.26KK281 pKa = 10.53KK282 pKa = 11.15YY283 pKa = 10.98NDD285 pKa = 3.2WGLNN289 pKa = 3.43
MM1 pKa = 7.38NIEE4 pKa = 4.18KK5 pKa = 10.76ALMNYY10 pKa = 9.87YY11 pKa = 10.37EE12 pKa = 4.53LTLASYY18 pKa = 10.82EE19 pKa = 4.21DD20 pKa = 3.58STNRR24 pKa = 11.84YY25 pKa = 8.16HH26 pKa = 7.19YY27 pKa = 10.36RR28 pKa = 11.84IVQHH32 pKa = 6.6LIKK35 pKa = 10.07GLKK38 pKa = 8.12YY39 pKa = 10.14IKK41 pKa = 10.09VKK43 pKa = 10.64KK44 pKa = 10.07LEE46 pKa = 4.33KK47 pKa = 10.06VDD49 pKa = 3.13ITIGYY54 pKa = 9.53KK55 pKa = 10.71LIDD58 pKa = 3.48YY59 pKa = 8.36LKK61 pKa = 10.2NHH63 pKa = 5.56THH65 pKa = 6.56NGNNSIKK72 pKa = 10.58KK73 pKa = 9.31IINYY77 pKa = 8.4LRR79 pKa = 11.84KK80 pKa = 9.74VMQHH84 pKa = 5.06YY85 pKa = 10.51RR86 pKa = 11.84ITTSIIDD93 pKa = 4.31LPHH96 pKa = 6.89LPNDD100 pKa = 3.77TKK102 pKa = 10.69PFEE105 pKa = 4.42RR106 pKa = 11.84FYY108 pKa = 11.19HH109 pKa = 7.2DD110 pKa = 4.35DD111 pKa = 3.86LEE113 pKa = 6.8LIMTYY118 pKa = 9.33TKK120 pKa = 10.97NLNSSKK126 pKa = 11.09NSITYY131 pKa = 10.16KK132 pKa = 10.61SFIRR136 pKa = 11.84LLLDD140 pKa = 3.25SGLRR144 pKa = 11.84VSEE147 pKa = 4.06ALNIKK152 pKa = 9.95ISDD155 pKa = 3.48MDD157 pKa = 4.08FKK159 pKa = 11.52NKK161 pKa = 9.59VIRR164 pKa = 11.84VLSSKK169 pKa = 7.56TRR171 pKa = 11.84KK172 pKa = 8.58QRR174 pKa = 11.84YY175 pKa = 8.83APFSSFSLKK184 pKa = 10.45YY185 pKa = 9.1IKK187 pKa = 10.21EE188 pKa = 4.28LIEE191 pKa = 3.98VNPKK195 pKa = 9.85RR196 pKa = 11.84DD197 pKa = 3.4YY198 pKa = 11.49LFYY201 pKa = 11.42NFIKK205 pKa = 10.58DD206 pKa = 3.33RR207 pKa = 11.84QTNKK211 pKa = 10.15NDD213 pKa = 2.59IKK215 pKa = 11.03LFYY218 pKa = 10.41KK219 pKa = 10.46RR220 pKa = 11.84LKK222 pKa = 10.12KK223 pKa = 9.99HH224 pKa = 6.2LNLEE228 pKa = 4.78RR229 pKa = 11.84IHH231 pKa = 5.08THH233 pKa = 5.76RR234 pKa = 11.84FRR236 pKa = 11.84KK237 pKa = 7.82TFASILIEE245 pKa = 4.23NGLNIDD251 pKa = 4.3DD252 pKa = 4.07LQKK255 pKa = 10.67IFDD258 pKa = 3.77HH259 pKa = 6.61SRR261 pKa = 11.84IEE263 pKa = 4.01TTIKK267 pKa = 10.3YY268 pKa = 8.37VQHH271 pKa = 5.74NEE273 pKa = 3.27KK274 pKa = 10.4RR275 pKa = 11.84ALQEE279 pKa = 3.82YY280 pKa = 10.26KK281 pKa = 10.53KK282 pKa = 11.15YY283 pKa = 10.98NDD285 pKa = 3.2WGLNN289 pKa = 3.43
Molecular weight: 34.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3273 |
64 |
738 |
233.8 |
26.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.286 ± 0.684 | 0.183 ± 0.115 |
5.805 ± 0.321 | 5.255 ± 0.637 |
4.919 ± 0.486 | 5.469 ± 0.626 |
1.558 ± 0.338 | 9.96 ± 0.618 |
7.241 ± 1.313 | 9.227 ± 0.553 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.955 ± 0.266 | 7.088 ± 0.819 |
2.903 ± 0.466 | 3.33 ± 0.34 |
3.422 ± 0.594 | 6.569 ± 0.465 |
6.386 ± 0.69 | 6.874 ± 0.827 |
1.436 ± 0.351 | 5.133 ± 0.509 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
