
Parendozoicomonas haliclonae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Endozoicomonadaceae; Parendozoicomonas
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
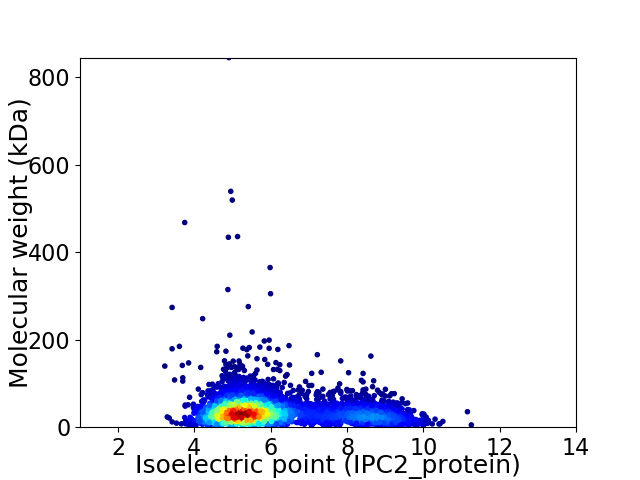
Virtual 2D-PAGE plot for 4695 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7AIY5|A0A1X7AIY5_9GAMM Hemin receptor OS=Parendozoicomonas haliclonae OX=1960125 GN=hemR PE=3 SV=1
MM1 pKa = 6.7FTVGEE6 pKa = 4.27VTGEE10 pKa = 4.04VRR12 pKa = 11.84AVAPDD17 pKa = 3.72GSSRR21 pKa = 11.84IVRR24 pKa = 11.84EE25 pKa = 3.99GDD27 pKa = 3.33SLLPGEE33 pKa = 4.3TLEE36 pKa = 4.91AEE38 pKa = 4.48EE39 pKa = 4.56GAQAILLFPDD49 pKa = 4.19GRR51 pKa = 11.84QLALPAGGTLTLPATTLSTQITDD74 pKa = 3.68LTEE77 pKa = 4.27SDD79 pKa = 3.81QSDD82 pKa = 3.77EE83 pKa = 4.89LKK85 pKa = 11.03ASDD88 pKa = 4.95DD89 pKa = 3.94SEE91 pKa = 4.67TIANSQADD99 pKa = 3.6GMLFPDD105 pKa = 4.02ISSSGFFEE113 pKa = 4.63SGPVSYY119 pKa = 10.89SADD122 pKa = 3.44SPNHH126 pKa = 5.35QEE128 pKa = 3.87NTPAASGQIGGGSSGPIAGLGSTPTLARR156 pKa = 11.84DD157 pKa = 3.62GSLGNSLDD165 pKa = 3.46SGVRR169 pKa = 11.84DD170 pKa = 3.03IGYY173 pKa = 7.52TSSRR177 pKa = 11.84GPVFSQSSDD186 pKa = 3.17PGISSGLSTVSSAGAIASSPSTAANSPIALSEE218 pKa = 4.0ALASGSGASSNTTPIISHH236 pKa = 5.61PTIVSSSATGGAVISSAQVQTAVSSAPATTSTPSTAPQQTSTQTITPASPSTPEE290 pKa = 3.74PAAQQATPTPTPEE303 pKa = 4.0PVVAASTTADD313 pKa = 3.04TGSVVFGQSLVQQSGLLSNDD333 pKa = 2.72QGTGITLSRR342 pKa = 11.84VGDD345 pKa = 3.65TDD347 pKa = 3.24ILEE350 pKa = 4.65TGSTTIQGQYY360 pKa = 10.86GSLSIQADD368 pKa = 3.06GMYY371 pKa = 10.1NYY373 pKa = 8.36TAASVDD379 pKa = 3.57LHH381 pKa = 8.53SDD383 pKa = 3.59MVAHH387 pKa = 6.95WAFNEE392 pKa = 4.54SVGSASVQDD401 pKa = 3.85SSIMDD406 pKa = 3.82SVTDD410 pKa = 3.63TGTLNNNASIVSGGRR425 pKa = 11.84TGNALQLDD433 pKa = 4.05GTGDD437 pKa = 3.83YY438 pKa = 11.39VSFADD443 pKa = 3.7SVEE446 pKa = 4.24INTPTSDD453 pKa = 2.56ISARR457 pKa = 11.84TISLFFQPDD466 pKa = 3.43SATSLSDD473 pKa = 3.04RR474 pKa = 11.84QVIYY478 pKa = 10.99EE479 pKa = 4.33EE480 pKa = 5.04GGTTNGFIIYY490 pKa = 9.3IQSNTLYY497 pKa = 10.26AGAYY501 pKa = 9.33SSSTGWTGHH510 pKa = 5.61YY511 pKa = 10.14FSTDD515 pKa = 2.42ISNLDD520 pKa = 3.25TSQWHH525 pKa = 6.55SIVLSLDD532 pKa = 2.95ASTNTMEE539 pKa = 4.13AWLDD543 pKa = 3.51GASIGEE549 pKa = 4.04ISNAQAVASHH559 pKa = 6.71ASDD562 pKa = 2.95IRR564 pKa = 11.84LGGSGDD570 pKa = 3.44GNLFHH575 pKa = 7.87DD576 pKa = 5.6GEE578 pKa = 4.46QTGAFNFNGKK588 pKa = 8.84IDD590 pKa = 3.65EE591 pKa = 4.15VRR593 pKa = 11.84VYY595 pKa = 11.05NRR597 pKa = 11.84ALNEE601 pKa = 4.03QEE603 pKa = 4.26VSLLHH608 pKa = 6.57SGNTSATEE616 pKa = 3.75IFNYY620 pKa = 8.83TATDD624 pKa = 3.55SAGGTSSSTLTLTVSTPSNISPVAVDD650 pKa = 4.24DD651 pKa = 4.29VLSVMSDD658 pKa = 2.59QSAAINSVAGQGLLGNDD675 pKa = 3.44RR676 pKa = 11.84DD677 pKa = 4.19AEE679 pKa = 4.43GDD681 pKa = 3.79SLTVTRR687 pKa = 11.84VGTTDD692 pKa = 3.01VAASGTTSITGTYY705 pKa = 7.08GTLVIAANGTYY716 pKa = 10.06SYY718 pKa = 11.09TPLNSAANGGTDD730 pKa = 2.99TFTYY734 pKa = 9.37TVSDD738 pKa = 3.6GTTTSTANLTVNVIADD754 pKa = 4.03GSANADD760 pKa = 3.57TLTVTEE766 pKa = 4.72SGFSASAVYY775 pKa = 9.83MLDD778 pKa = 3.18EE779 pKa = 4.93TGGQDD784 pKa = 3.36YY785 pKa = 10.61LAVVDD790 pKa = 4.12TEE792 pKa = 4.61SGVSHH797 pKa = 7.29RR798 pKa = 11.84LGAITGSSASRR809 pKa = 11.84KK810 pKa = 6.58TLAMGPDD817 pKa = 3.21GTLYY821 pKa = 9.74VTDD824 pKa = 3.71STNLYY829 pKa = 8.81TLDD832 pKa = 4.58PSTQALTLVGAHH844 pKa = 6.13GVTDD848 pKa = 3.5SMSFIGLTVAPDD860 pKa = 3.54GTIYY864 pKa = 10.06AAKK867 pKa = 9.85YY868 pKa = 9.48DD869 pKa = 3.61GHH871 pKa = 8.25IYY873 pKa = 9.34TLNPSTGAATDD884 pKa = 3.71LGGVSGFEE892 pKa = 4.23NNWGDD897 pKa = 4.71LVWHH901 pKa = 7.25DD902 pKa = 3.51GALYY906 pKa = 7.98TQAYY910 pKa = 7.0TYY912 pKa = 11.18TNGQSIYY919 pKa = 9.62QFVRR923 pKa = 11.84IEE925 pKa = 3.95PSNSGFTATPLPNASDD941 pKa = 3.62AGSLHH946 pKa = 7.2GITSVNGEE954 pKa = 3.88LRR956 pKa = 11.84GVVWSYY962 pKa = 11.2TGGEE966 pKa = 4.41TVTIDD971 pKa = 3.34TTTGLVSSRR980 pKa = 11.84APVTGAAKK988 pKa = 10.21IEE990 pKa = 4.44DD991 pKa = 4.15SASSGTSMAGNVLTNDD1007 pKa = 3.19TGTTGVTTVALPDD1020 pKa = 3.94GTSSTVSNGGATIQGTYY1037 pKa = 8.63GTLIMQTDD1045 pKa = 3.86GSYY1048 pKa = 10.23IYY1050 pKa = 10.38TLDD1053 pKa = 3.62NSLEE1057 pKa = 4.1ATNNLASEE1065 pKa = 4.4EE1066 pKa = 4.33TGTDD1070 pKa = 2.63RR1071 pKa = 11.84FVYY1074 pKa = 9.17TASTTGGSTQSILTVFVNGSSEE1096 pKa = 4.57VVTTDD1101 pKa = 2.64AATFSNYY1108 pKa = 9.5NVMTSVDD1115 pKa = 3.86ASSSTIPIDD1124 pKa = 4.05FTITTQNDD1132 pKa = 3.04SDD1134 pKa = 4.81KK1135 pKa = 9.86ITEE1138 pKa = 4.06IVISGVNGSTFNVPSVLTDD1157 pKa = 2.94NGTITGNTSNSSEE1170 pKa = 4.35FVWRR1174 pKa = 11.84ATPGTNAGLDD1184 pKa = 3.67SFDD1187 pKa = 3.86AASAGLTLATINGSQAWGQHH1207 pKa = 4.76MGISATVSEE1216 pKa = 4.59YY1217 pKa = 11.22DD1218 pKa = 3.59SNGATTSSWTSTSAIHH1234 pKa = 5.41TTLAGIDD1241 pKa = 3.5HH1242 pKa = 6.81TRR1244 pKa = 11.84TGTSGDD1250 pKa = 3.63DD1251 pKa = 3.52TLTGSSDD1258 pKa = 2.45TWTNIDD1264 pKa = 4.08EE1265 pKa = 5.23FIGGAGNDD1273 pKa = 3.58TMTGGSGSDD1282 pKa = 3.15FYY1284 pKa = 11.55VWRR1287 pKa = 11.84GADD1290 pKa = 3.31VGTSSSPAIDD1300 pKa = 4.2TITDD1304 pKa = 4.0FSAGRR1309 pKa = 11.84LGDD1312 pKa = 3.74VLDD1315 pKa = 4.56LRR1317 pKa = 11.84EE1318 pKa = 4.8LLPDD1322 pKa = 3.67TAEE1325 pKa = 4.05STLDD1329 pKa = 3.37EE1330 pKa = 4.4FLSFSFADD1338 pKa = 3.79GNTTIGVSTTAGGPVVQNIVLDD1360 pKa = 4.03NVDD1363 pKa = 4.19LSSSFGTTDD1372 pKa = 2.93VTQLTNQLTDD1382 pKa = 3.06NGNLLTT1388 pKa = 4.76
MM1 pKa = 6.7FTVGEE6 pKa = 4.27VTGEE10 pKa = 4.04VRR12 pKa = 11.84AVAPDD17 pKa = 3.72GSSRR21 pKa = 11.84IVRR24 pKa = 11.84EE25 pKa = 3.99GDD27 pKa = 3.33SLLPGEE33 pKa = 4.3TLEE36 pKa = 4.91AEE38 pKa = 4.48EE39 pKa = 4.56GAQAILLFPDD49 pKa = 4.19GRR51 pKa = 11.84QLALPAGGTLTLPATTLSTQITDD74 pKa = 3.68LTEE77 pKa = 4.27SDD79 pKa = 3.81QSDD82 pKa = 3.77EE83 pKa = 4.89LKK85 pKa = 11.03ASDD88 pKa = 4.95DD89 pKa = 3.94SEE91 pKa = 4.67TIANSQADD99 pKa = 3.6GMLFPDD105 pKa = 4.02ISSSGFFEE113 pKa = 4.63SGPVSYY119 pKa = 10.89SADD122 pKa = 3.44SPNHH126 pKa = 5.35QEE128 pKa = 3.87NTPAASGQIGGGSSGPIAGLGSTPTLARR156 pKa = 11.84DD157 pKa = 3.62GSLGNSLDD165 pKa = 3.46SGVRR169 pKa = 11.84DD170 pKa = 3.03IGYY173 pKa = 7.52TSSRR177 pKa = 11.84GPVFSQSSDD186 pKa = 3.17PGISSGLSTVSSAGAIASSPSTAANSPIALSEE218 pKa = 4.0ALASGSGASSNTTPIISHH236 pKa = 5.61PTIVSSSATGGAVISSAQVQTAVSSAPATTSTPSTAPQQTSTQTITPASPSTPEE290 pKa = 3.74PAAQQATPTPTPEE303 pKa = 4.0PVVAASTTADD313 pKa = 3.04TGSVVFGQSLVQQSGLLSNDD333 pKa = 2.72QGTGITLSRR342 pKa = 11.84VGDD345 pKa = 3.65TDD347 pKa = 3.24ILEE350 pKa = 4.65TGSTTIQGQYY360 pKa = 10.86GSLSIQADD368 pKa = 3.06GMYY371 pKa = 10.1NYY373 pKa = 8.36TAASVDD379 pKa = 3.57LHH381 pKa = 8.53SDD383 pKa = 3.59MVAHH387 pKa = 6.95WAFNEE392 pKa = 4.54SVGSASVQDD401 pKa = 3.85SSIMDD406 pKa = 3.82SVTDD410 pKa = 3.63TGTLNNNASIVSGGRR425 pKa = 11.84TGNALQLDD433 pKa = 4.05GTGDD437 pKa = 3.83YY438 pKa = 11.39VSFADD443 pKa = 3.7SVEE446 pKa = 4.24INTPTSDD453 pKa = 2.56ISARR457 pKa = 11.84TISLFFQPDD466 pKa = 3.43SATSLSDD473 pKa = 3.04RR474 pKa = 11.84QVIYY478 pKa = 10.99EE479 pKa = 4.33EE480 pKa = 5.04GGTTNGFIIYY490 pKa = 9.3IQSNTLYY497 pKa = 10.26AGAYY501 pKa = 9.33SSSTGWTGHH510 pKa = 5.61YY511 pKa = 10.14FSTDD515 pKa = 2.42ISNLDD520 pKa = 3.25TSQWHH525 pKa = 6.55SIVLSLDD532 pKa = 2.95ASTNTMEE539 pKa = 4.13AWLDD543 pKa = 3.51GASIGEE549 pKa = 4.04ISNAQAVASHH559 pKa = 6.71ASDD562 pKa = 2.95IRR564 pKa = 11.84LGGSGDD570 pKa = 3.44GNLFHH575 pKa = 7.87DD576 pKa = 5.6GEE578 pKa = 4.46QTGAFNFNGKK588 pKa = 8.84IDD590 pKa = 3.65EE591 pKa = 4.15VRR593 pKa = 11.84VYY595 pKa = 11.05NRR597 pKa = 11.84ALNEE601 pKa = 4.03QEE603 pKa = 4.26VSLLHH608 pKa = 6.57SGNTSATEE616 pKa = 3.75IFNYY620 pKa = 8.83TATDD624 pKa = 3.55SAGGTSSSTLTLTVSTPSNISPVAVDD650 pKa = 4.24DD651 pKa = 4.29VLSVMSDD658 pKa = 2.59QSAAINSVAGQGLLGNDD675 pKa = 3.44RR676 pKa = 11.84DD677 pKa = 4.19AEE679 pKa = 4.43GDD681 pKa = 3.79SLTVTRR687 pKa = 11.84VGTTDD692 pKa = 3.01VAASGTTSITGTYY705 pKa = 7.08GTLVIAANGTYY716 pKa = 10.06SYY718 pKa = 11.09TPLNSAANGGTDD730 pKa = 2.99TFTYY734 pKa = 9.37TVSDD738 pKa = 3.6GTTTSTANLTVNVIADD754 pKa = 4.03GSANADD760 pKa = 3.57TLTVTEE766 pKa = 4.72SGFSASAVYY775 pKa = 9.83MLDD778 pKa = 3.18EE779 pKa = 4.93TGGQDD784 pKa = 3.36YY785 pKa = 10.61LAVVDD790 pKa = 4.12TEE792 pKa = 4.61SGVSHH797 pKa = 7.29RR798 pKa = 11.84LGAITGSSASRR809 pKa = 11.84KK810 pKa = 6.58TLAMGPDD817 pKa = 3.21GTLYY821 pKa = 9.74VTDD824 pKa = 3.71STNLYY829 pKa = 8.81TLDD832 pKa = 4.58PSTQALTLVGAHH844 pKa = 6.13GVTDD848 pKa = 3.5SMSFIGLTVAPDD860 pKa = 3.54GTIYY864 pKa = 10.06AAKK867 pKa = 9.85YY868 pKa = 9.48DD869 pKa = 3.61GHH871 pKa = 8.25IYY873 pKa = 9.34TLNPSTGAATDD884 pKa = 3.71LGGVSGFEE892 pKa = 4.23NNWGDD897 pKa = 4.71LVWHH901 pKa = 7.25DD902 pKa = 3.51GALYY906 pKa = 7.98TQAYY910 pKa = 7.0TYY912 pKa = 11.18TNGQSIYY919 pKa = 9.62QFVRR923 pKa = 11.84IEE925 pKa = 3.95PSNSGFTATPLPNASDD941 pKa = 3.62AGSLHH946 pKa = 7.2GITSVNGEE954 pKa = 3.88LRR956 pKa = 11.84GVVWSYY962 pKa = 11.2TGGEE966 pKa = 4.41TVTIDD971 pKa = 3.34TTTGLVSSRR980 pKa = 11.84APVTGAAKK988 pKa = 10.21IEE990 pKa = 4.44DD991 pKa = 4.15SASSGTSMAGNVLTNDD1007 pKa = 3.19TGTTGVTTVALPDD1020 pKa = 3.94GTSSTVSNGGATIQGTYY1037 pKa = 8.63GTLIMQTDD1045 pKa = 3.86GSYY1048 pKa = 10.23IYY1050 pKa = 10.38TLDD1053 pKa = 3.62NSLEE1057 pKa = 4.1ATNNLASEE1065 pKa = 4.4EE1066 pKa = 4.33TGTDD1070 pKa = 2.63RR1071 pKa = 11.84FVYY1074 pKa = 9.17TASTTGGSTQSILTVFVNGSSEE1096 pKa = 4.57VVTTDD1101 pKa = 2.64AATFSNYY1108 pKa = 9.5NVMTSVDD1115 pKa = 3.86ASSSTIPIDD1124 pKa = 4.05FTITTQNDD1132 pKa = 3.04SDD1134 pKa = 4.81KK1135 pKa = 9.86ITEE1138 pKa = 4.06IVISGVNGSTFNVPSVLTDD1157 pKa = 2.94NGTITGNTSNSSEE1170 pKa = 4.35FVWRR1174 pKa = 11.84ATPGTNAGLDD1184 pKa = 3.67SFDD1187 pKa = 3.86AASAGLTLATINGSQAWGQHH1207 pKa = 4.76MGISATVSEE1216 pKa = 4.59YY1217 pKa = 11.22DD1218 pKa = 3.59SNGATTSSWTSTSAIHH1234 pKa = 5.41TTLAGIDD1241 pKa = 3.5HH1242 pKa = 6.81TRR1244 pKa = 11.84TGTSGDD1250 pKa = 3.63DD1251 pKa = 3.52TLTGSSDD1258 pKa = 2.45TWTNIDD1264 pKa = 4.08EE1265 pKa = 5.23FIGGAGNDD1273 pKa = 3.58TMTGGSGSDD1282 pKa = 3.15FYY1284 pKa = 11.55VWRR1287 pKa = 11.84GADD1290 pKa = 3.31VGTSSSPAIDD1300 pKa = 4.2TITDD1304 pKa = 4.0FSAGRR1309 pKa = 11.84LGDD1312 pKa = 3.74VLDD1315 pKa = 4.56LRR1317 pKa = 11.84EE1318 pKa = 4.8LLPDD1322 pKa = 3.67TAEE1325 pKa = 4.05STLDD1329 pKa = 3.37EE1330 pKa = 4.4FLSFSFADD1338 pKa = 3.79GNTTIGVSTTAGGPVVQNIVLDD1360 pKa = 4.03NVDD1363 pKa = 4.19LSSSFGTTDD1372 pKa = 2.93VTQLTNQLTDD1382 pKa = 3.06NGNLLTT1388 pKa = 4.76
Molecular weight: 141.22 kDa
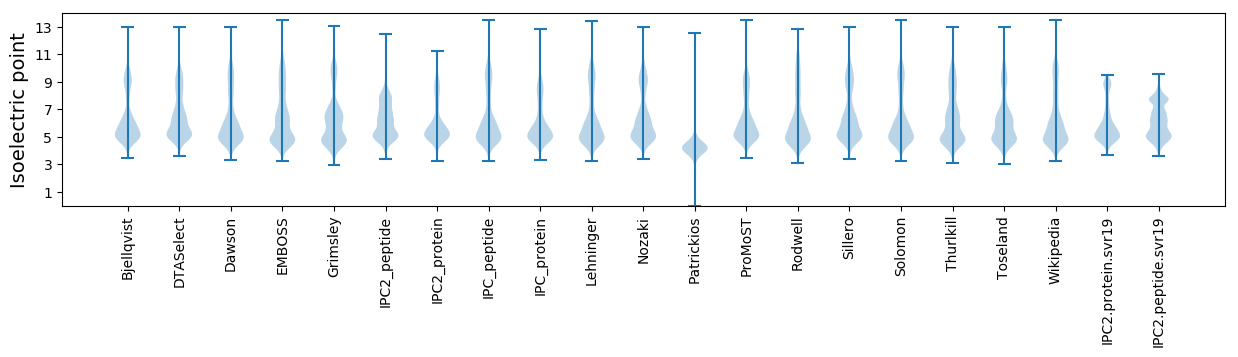
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7AJD2|A0A1X7AJD2_9GAMM Histone H1-like protein HC2 OS=Parendozoicomonas haliclonae OX=1960125 GN=hctB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84AVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84AVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1626144 |
29 |
7743 |
346.4 |
38.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.909 ± 0.035 | 1.106 ± 0.014 |
5.6 ± 0.04 | 6.361 ± 0.037 |
3.781 ± 0.025 | 7.297 ± 0.034 |
2.258 ± 0.018 | 5.612 ± 0.029 |
4.605 ± 0.038 | 10.484 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.702 ± 0.022 | 3.838 ± 0.028 |
4.551 ± 0.031 | 4.556 ± 0.033 |
5.338 ± 0.03 | 6.68 ± 0.037 |
5.511 ± 0.033 | 6.707 ± 0.031 |
1.295 ± 0.013 | 2.807 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
