
Oscillibacter sp. PC13
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Oscillibacter; unclassified Oscillibacter
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
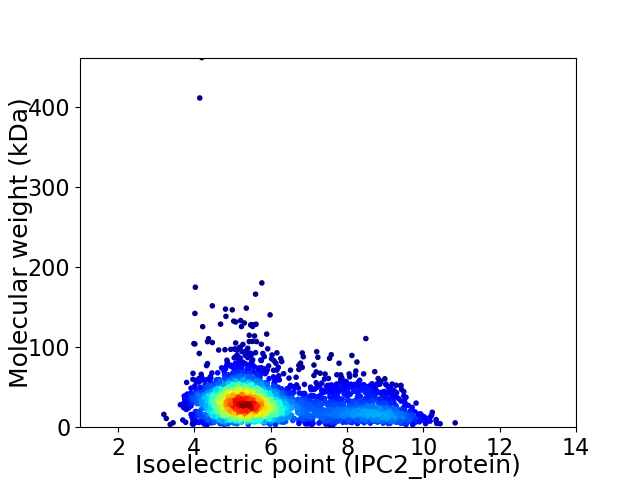
Virtual 2D-PAGE plot for 2711 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I5SCQ1|A0A1I5SCQ1_9FIRM Ferrous iron transport protein B OS=Oscillibacter sp. PC13 OX=1855299 GN=SAMN05216343_11216 PE=3 SV=1
MM1 pKa = 7.53KK2 pKa = 10.51KK3 pKa = 10.26FFALLLTLACVLSLAACGSEE23 pKa = 3.97AAPTEE28 pKa = 4.38TPDD31 pKa = 3.22NQTNEE36 pKa = 4.44DD37 pKa = 3.89PADD40 pKa = 3.58TAFTTVEE47 pKa = 3.85EE48 pKa = 5.1GKK50 pKa = 10.33LHH52 pKa = 6.13MATNAAFPPYY62 pKa = 10.98EE63 pKa = 4.16MTTDD67 pKa = 3.08AGGFEE72 pKa = 5.05GIDD75 pKa = 3.27VDD77 pKa = 4.16VAAAIAEE84 pKa = 4.24KK85 pKa = 10.65LGLEE89 pKa = 4.17LVVDD93 pKa = 4.49DD94 pKa = 4.76MDD96 pKa = 4.27FNSVITAVQQGSSDD110 pKa = 2.83IAMAGLTVNEE120 pKa = 3.94EE121 pKa = 3.93RR122 pKa = 11.84KK123 pKa = 10.16QNVDD127 pKa = 3.76FTASYY132 pKa = 8.43ATGVQVVIVKK142 pKa = 10.34AGSDD146 pKa = 3.07ITLDD150 pKa = 3.59NLGDD154 pKa = 3.77QMIGTQEE161 pKa = 3.92STTGYY166 pKa = 9.63IYY168 pKa = 10.94CADD171 pKa = 4.05EE172 pKa = 4.35FGEE175 pKa = 4.3DD176 pKa = 3.69HH177 pKa = 7.12VIAYY181 pKa = 7.51TNGATAVQSLLAGSVDD197 pKa = 3.77CVVIDD202 pKa = 4.2NEE204 pKa = 4.05PAKK207 pKa = 10.88AFVAANPDD215 pKa = 3.55LTILDD220 pKa = 3.54TEE222 pKa = 4.67YY223 pKa = 11.17AVEE226 pKa = 4.88DD227 pKa = 3.86YY228 pKa = 11.16AIGVSKK234 pKa = 10.58EE235 pKa = 3.73NSALLEE241 pKa = 4.34AVDD244 pKa = 4.4TALQDD249 pKa = 4.53LIADD253 pKa = 4.12GTVQSIVDD261 pKa = 3.46KK262 pKa = 11.13YY263 pKa = 9.58ITAEE267 pKa = 3.75
MM1 pKa = 7.53KK2 pKa = 10.51KK3 pKa = 10.26FFALLLTLACVLSLAACGSEE23 pKa = 3.97AAPTEE28 pKa = 4.38TPDD31 pKa = 3.22NQTNEE36 pKa = 4.44DD37 pKa = 3.89PADD40 pKa = 3.58TAFTTVEE47 pKa = 3.85EE48 pKa = 5.1GKK50 pKa = 10.33LHH52 pKa = 6.13MATNAAFPPYY62 pKa = 10.98EE63 pKa = 4.16MTTDD67 pKa = 3.08AGGFEE72 pKa = 5.05GIDD75 pKa = 3.27VDD77 pKa = 4.16VAAAIAEE84 pKa = 4.24KK85 pKa = 10.65LGLEE89 pKa = 4.17LVVDD93 pKa = 4.49DD94 pKa = 4.76MDD96 pKa = 4.27FNSVITAVQQGSSDD110 pKa = 2.83IAMAGLTVNEE120 pKa = 3.94EE121 pKa = 3.93RR122 pKa = 11.84KK123 pKa = 10.16QNVDD127 pKa = 3.76FTASYY132 pKa = 8.43ATGVQVVIVKK142 pKa = 10.34AGSDD146 pKa = 3.07ITLDD150 pKa = 3.59NLGDD154 pKa = 3.77QMIGTQEE161 pKa = 3.92STTGYY166 pKa = 9.63IYY168 pKa = 10.94CADD171 pKa = 4.05EE172 pKa = 4.35FGEE175 pKa = 4.3DD176 pKa = 3.69HH177 pKa = 7.12VIAYY181 pKa = 7.51TNGATAVQSLLAGSVDD197 pKa = 3.77CVVIDD202 pKa = 4.2NEE204 pKa = 4.05PAKK207 pKa = 10.88AFVAANPDD215 pKa = 3.55LTILDD220 pKa = 3.54TEE222 pKa = 4.67YY223 pKa = 11.17AVEE226 pKa = 4.88DD227 pKa = 3.86YY228 pKa = 11.16AIGVSKK234 pKa = 10.58EE235 pKa = 3.73NSALLEE241 pKa = 4.34AVDD244 pKa = 4.4TALQDD249 pKa = 4.53LIADD253 pKa = 4.12GTVQSIVDD261 pKa = 3.46KK262 pKa = 11.13YY263 pKa = 9.58ITAEE267 pKa = 3.75
Molecular weight: 28.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I5U3E9|A0A1I5U3E9_9FIRM 30S ribosomal protein S20 OS=Oscillibacter sp. PC13 OX=1855299 GN=rpsT PE=3 SV=1
MM1 pKa = 7.51AFDD4 pKa = 3.87RR5 pKa = 11.84EE6 pKa = 4.29ARR8 pKa = 11.84PARR11 pKa = 11.84PMRR14 pKa = 11.84GKK16 pKa = 10.06RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.1VCQFCADD26 pKa = 3.26KK27 pKa = 11.23CEE29 pKa = 4.7SIDD32 pKa = 3.69YY33 pKa = 10.71KK34 pKa = 11.09DD35 pKa = 3.24AAKK38 pKa = 10.4LRR40 pKa = 11.84RR41 pKa = 11.84FVSEE45 pKa = 3.77RR46 pKa = 11.84SKK48 pKa = 10.62ILPRR52 pKa = 11.84RR53 pKa = 11.84TTGTCAMHH61 pKa = 6.16QRR63 pKa = 11.84QLTEE67 pKa = 4.31AIKK70 pKa = 10.35RR71 pKa = 11.84ARR73 pKa = 11.84QIALLPYY80 pKa = 9.27VTDD83 pKa = 3.53
MM1 pKa = 7.51AFDD4 pKa = 3.87RR5 pKa = 11.84EE6 pKa = 4.29ARR8 pKa = 11.84PARR11 pKa = 11.84PMRR14 pKa = 11.84GKK16 pKa = 10.06RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.1VCQFCADD26 pKa = 3.26KK27 pKa = 11.23CEE29 pKa = 4.7SIDD32 pKa = 3.69YY33 pKa = 10.71KK34 pKa = 11.09DD35 pKa = 3.24AAKK38 pKa = 10.4LRR40 pKa = 11.84RR41 pKa = 11.84FVSEE45 pKa = 3.77RR46 pKa = 11.84SKK48 pKa = 10.62ILPRR52 pKa = 11.84RR53 pKa = 11.84TTGTCAMHH61 pKa = 6.16QRR63 pKa = 11.84QLTEE67 pKa = 4.31AIKK70 pKa = 10.35RR71 pKa = 11.84ARR73 pKa = 11.84QIALLPYY80 pKa = 9.27VTDD83 pKa = 3.53
Molecular weight: 9.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
811945 |
25 |
4336 |
299.5 |
33.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.837 ± 0.058 | 1.732 ± 0.025 |
5.423 ± 0.049 | 6.702 ± 0.05 |
3.829 ± 0.032 | 7.915 ± 0.046 |
1.869 ± 0.024 | 5.803 ± 0.045 |
4.799 ± 0.049 | 9.959 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.939 ± 0.03 | 3.333 ± 0.03 |
4.098 ± 0.028 | 3.416 ± 0.026 |
5.408 ± 0.054 | 5.566 ± 0.043 |
5.68 ± 0.069 | 7.201 ± 0.04 |
1.044 ± 0.019 | 3.447 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
