
Pelagicola litoralis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Pelagicola
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
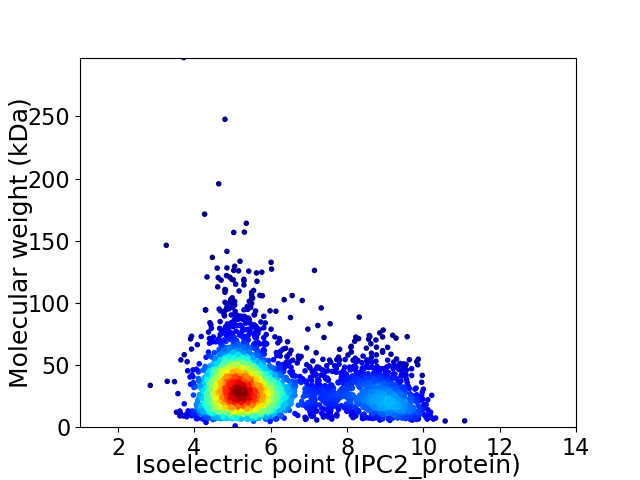
Virtual 2D-PAGE plot for 3367 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U7MRW7|A0A4U7MRW7_9RHOB Uncharacterized protein OS=Pelagicola litoralis OX=420403 GN=FAP39_17065 PE=4 SV=1
GG1 pKa = 6.82ATQQVTVPVTVTDD14 pKa = 3.6STGATDD20 pKa = 3.63TEE22 pKa = 4.24NLVITVTGTNDD33 pKa = 2.94GPAVSGPVTLPGGTEE48 pKa = 3.91DD49 pKa = 3.44TSVQITAAQLLEE61 pKa = 4.4HH62 pKa = 6.59ATDD65 pKa = 4.66ADD67 pKa = 3.76TGDD70 pKa = 3.55QLAVTGLTASHH81 pKa = 5.91GTITGDD87 pKa = 3.08AATGFTFTPDD97 pKa = 2.89ANYY100 pKa = 10.54NGPVSLSYY108 pKa = 10.47TVTDD112 pKa = 3.3GHH114 pKa = 6.54GGSVAQTASLTLGAVGDD131 pKa = 3.79PAIIGGVDD139 pKa = 3.05TGDD142 pKa = 3.37VTEE145 pKa = 4.84NKK147 pKa = 9.95AGQDD151 pKa = 3.45MSPDD155 pKa = 3.41YY156 pKa = 10.13AQPGMATLGRR166 pKa = 11.84AQLYY170 pKa = 10.83ADD172 pKa = 4.11GQLTILDD179 pKa = 4.57PDD181 pKa = 3.53TGEE184 pKa = 4.05NQFDD188 pKa = 3.9TRR190 pKa = 11.84GSMGMNYY197 pKa = 10.11SGTYY201 pKa = 10.01GDD203 pKa = 5.35LRR205 pKa = 11.84LNSDD209 pKa = 4.2GSWHH213 pKa = 6.26YY214 pKa = 11.19YY215 pKa = 9.58GDD217 pKa = 3.92AGHH220 pKa = 6.8IRR222 pKa = 11.84VTGGRR227 pKa = 11.84TTTRR231 pKa = 11.84GTAIDD236 pKa = 3.64QLGEE240 pKa = 4.22GQTLTDD246 pKa = 4.46TITVYY251 pKa = 11.07SKK253 pKa = 11.02DD254 pKa = 3.4GTAHH258 pKa = 7.58DD259 pKa = 3.75ITITIHH265 pKa = 6.11GSNDD269 pKa = 2.92RR270 pKa = 11.84PYY272 pKa = 10.65CSSEE276 pKa = 3.92VALTPGTEE284 pKa = 4.22DD285 pKa = 3.16TAQTLTKK292 pKa = 10.45AQLLANTVEE301 pKa = 4.01VDD303 pKa = 3.29ANDD306 pKa = 3.71AGKK309 pKa = 10.77LSIAGLRR316 pKa = 11.84ADD318 pKa = 4.1HH319 pKa = 7.17GSIRR323 pKa = 11.84DD324 pKa = 3.92NGDD327 pKa = 2.64GTYY330 pKa = 10.28TFTPARR336 pKa = 11.84DD337 pKa = 3.62YY338 pKa = 11.55NGAVHH343 pKa = 6.2FTYY346 pKa = 10.23DD347 pKa = 3.07VKK349 pKa = 10.92DD350 pKa = 3.26AHH352 pKa = 6.92GGVTHH357 pKa = 7.18TGATTTLAAVSDD369 pKa = 3.81AAIIGGTDD377 pKa = 2.98TGSATEE383 pKa = 3.87DD384 pKa = 3.46HH385 pKa = 5.47VTGRR389 pKa = 11.84IVISGLLTVSDD400 pKa = 4.52PDD402 pKa = 4.13GPSQEE407 pKa = 4.2HH408 pKa = 5.16FQYY411 pKa = 11.05SQFGEE416 pKa = 4.24HH417 pKa = 7.14AISDD421 pKa = 3.95PFGGNLHH428 pKa = 6.76IDD430 pKa = 3.73SAGSWSYY437 pKa = 9.36VTDD440 pKa = 3.5NSNPAVQQLAAGQEE454 pKa = 4.0GHH456 pKa = 5.06ATYY459 pKa = 10.36RR460 pKa = 11.84VRR462 pKa = 11.84SSDD465 pKa = 3.16GTTHH469 pKa = 6.83QIQITIHH476 pKa = 5.47GTNDD480 pKa = 3.23APVLSASTASATEE493 pKa = 4.6DD494 pKa = 3.72GQSVTGQVSGTDD506 pKa = 3.2VDD508 pKa = 3.74TGDD511 pKa = 3.66ALAYY515 pKa = 10.19SLGSAAPAGFTLNPDD530 pKa = 3.81GSWSFDD536 pKa = 3.69PADD539 pKa = 3.58AAYY542 pKa = 10.19QHH544 pKa = 6.84LAAGATQQVTVPVTVTDD561 pKa = 3.6STGATDD567 pKa = 3.59TQNLVITVTGTNDD580 pKa = 3.06GPTVSGPVTLPGGTEE595 pKa = 3.91DD596 pKa = 3.44TSVQITAAQLLAHH609 pKa = 6.49ATDD612 pKa = 4.02IDD614 pKa = 3.89TGDD617 pKa = 3.21TT618 pKa = 3.49
GG1 pKa = 6.82ATQQVTVPVTVTDD14 pKa = 3.6STGATDD20 pKa = 3.63TEE22 pKa = 4.24NLVITVTGTNDD33 pKa = 2.94GPAVSGPVTLPGGTEE48 pKa = 3.91DD49 pKa = 3.44TSVQITAAQLLEE61 pKa = 4.4HH62 pKa = 6.59ATDD65 pKa = 4.66ADD67 pKa = 3.76TGDD70 pKa = 3.55QLAVTGLTASHH81 pKa = 5.91GTITGDD87 pKa = 3.08AATGFTFTPDD97 pKa = 2.89ANYY100 pKa = 10.54NGPVSLSYY108 pKa = 10.47TVTDD112 pKa = 3.3GHH114 pKa = 6.54GGSVAQTASLTLGAVGDD131 pKa = 3.79PAIIGGVDD139 pKa = 3.05TGDD142 pKa = 3.37VTEE145 pKa = 4.84NKK147 pKa = 9.95AGQDD151 pKa = 3.45MSPDD155 pKa = 3.41YY156 pKa = 10.13AQPGMATLGRR166 pKa = 11.84AQLYY170 pKa = 10.83ADD172 pKa = 4.11GQLTILDD179 pKa = 4.57PDD181 pKa = 3.53TGEE184 pKa = 4.05NQFDD188 pKa = 3.9TRR190 pKa = 11.84GSMGMNYY197 pKa = 10.11SGTYY201 pKa = 10.01GDD203 pKa = 5.35LRR205 pKa = 11.84LNSDD209 pKa = 4.2GSWHH213 pKa = 6.26YY214 pKa = 11.19YY215 pKa = 9.58GDD217 pKa = 3.92AGHH220 pKa = 6.8IRR222 pKa = 11.84VTGGRR227 pKa = 11.84TTTRR231 pKa = 11.84GTAIDD236 pKa = 3.64QLGEE240 pKa = 4.22GQTLTDD246 pKa = 4.46TITVYY251 pKa = 11.07SKK253 pKa = 11.02DD254 pKa = 3.4GTAHH258 pKa = 7.58DD259 pKa = 3.75ITITIHH265 pKa = 6.11GSNDD269 pKa = 2.92RR270 pKa = 11.84PYY272 pKa = 10.65CSSEE276 pKa = 3.92VALTPGTEE284 pKa = 4.22DD285 pKa = 3.16TAQTLTKK292 pKa = 10.45AQLLANTVEE301 pKa = 4.01VDD303 pKa = 3.29ANDD306 pKa = 3.71AGKK309 pKa = 10.77LSIAGLRR316 pKa = 11.84ADD318 pKa = 4.1HH319 pKa = 7.17GSIRR323 pKa = 11.84DD324 pKa = 3.92NGDD327 pKa = 2.64GTYY330 pKa = 10.28TFTPARR336 pKa = 11.84DD337 pKa = 3.62YY338 pKa = 11.55NGAVHH343 pKa = 6.2FTYY346 pKa = 10.23DD347 pKa = 3.07VKK349 pKa = 10.92DD350 pKa = 3.26AHH352 pKa = 6.92GGVTHH357 pKa = 7.18TGATTTLAAVSDD369 pKa = 3.81AAIIGGTDD377 pKa = 2.98TGSATEE383 pKa = 3.87DD384 pKa = 3.46HH385 pKa = 5.47VTGRR389 pKa = 11.84IVISGLLTVSDD400 pKa = 4.52PDD402 pKa = 4.13GPSQEE407 pKa = 4.2HH408 pKa = 5.16FQYY411 pKa = 11.05SQFGEE416 pKa = 4.24HH417 pKa = 7.14AISDD421 pKa = 3.95PFGGNLHH428 pKa = 6.76IDD430 pKa = 3.73SAGSWSYY437 pKa = 9.36VTDD440 pKa = 3.5NSNPAVQQLAAGQEE454 pKa = 4.0GHH456 pKa = 5.06ATYY459 pKa = 10.36RR460 pKa = 11.84VRR462 pKa = 11.84SSDD465 pKa = 3.16GTTHH469 pKa = 6.83QIQITIHH476 pKa = 5.47GTNDD480 pKa = 3.23APVLSASTASATEE493 pKa = 4.6DD494 pKa = 3.72GQSVTGQVSGTDD506 pKa = 3.2VDD508 pKa = 3.74TGDD511 pKa = 3.66ALAYY515 pKa = 10.19SLGSAAPAGFTLNPDD530 pKa = 3.81GSWSFDD536 pKa = 3.69PADD539 pKa = 3.58AAYY542 pKa = 10.19QHH544 pKa = 6.84LAAGATQQVTVPVTVTDD561 pKa = 3.6STGATDD567 pKa = 3.59TQNLVITVTGTNDD580 pKa = 3.06GPTVSGPVTLPGGTEE595 pKa = 3.91DD596 pKa = 3.44TSVQITAAQLLAHH609 pKa = 6.49ATDD612 pKa = 4.02IDD614 pKa = 3.89TGDD617 pKa = 3.21TT618 pKa = 3.49
Molecular weight: 62.85 kDa
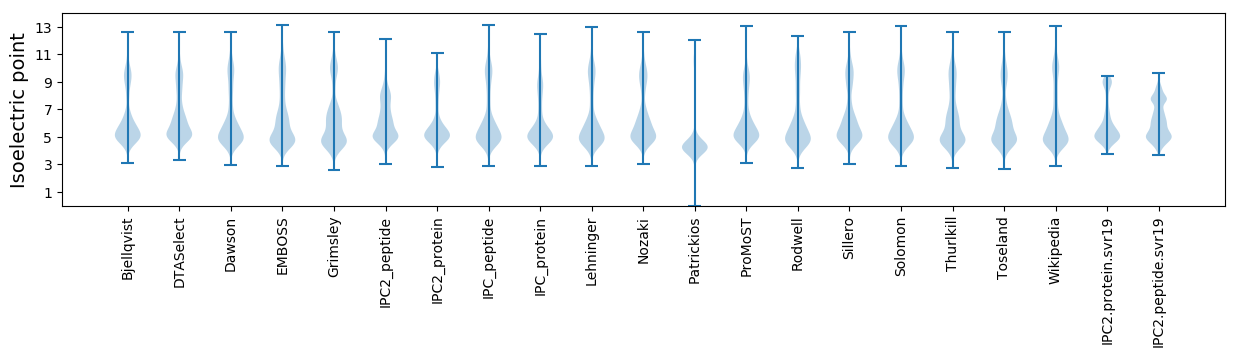
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4U7N8S8|A0A4U7N8S8_9RHOB ABC transporter ATP-binding protein OS=Pelagicola litoralis OX=420403 GN=FAP39_00465 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.34SLSAA44 pKa = 3.83
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.34SLSAA44 pKa = 3.83
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1051415 |
8 |
2823 |
312.3 |
34.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.347 ± 0.052 | 0.931 ± 0.013 |
6.286 ± 0.042 | 5.626 ± 0.04 |
3.852 ± 0.023 | 8.323 ± 0.041 |
2.208 ± 0.02 | 5.403 ± 0.029 |
3.714 ± 0.033 | 9.757 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.941 ± 0.022 | 2.883 ± 0.023 |
4.799 ± 0.026 | 3.565 ± 0.024 |
6.043 ± 0.038 | 5.599 ± 0.028 |
5.657 ± 0.028 | 7.339 ± 0.033 |
1.389 ± 0.017 | 2.338 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
