
Pelagibacter phage HTVC025P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; unclassified Autographiviridae
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
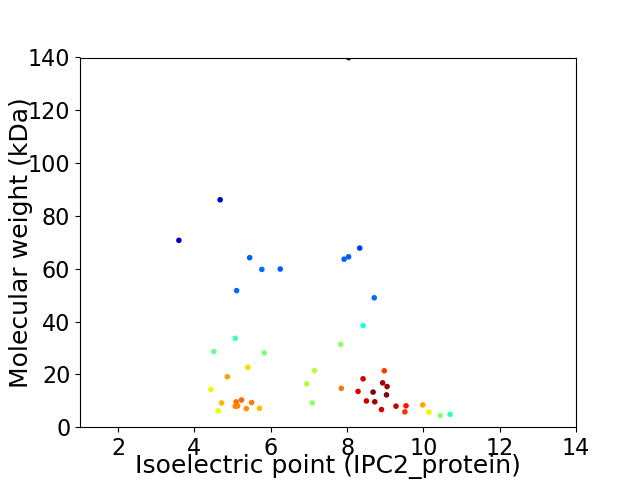
Virtual 2D-PAGE plot for 49 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1NU62|A0A4Y1NU62_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC025P OX=2259657 GN=P025_gp38 PE=4 SV=1
MM1 pKa = 7.49SFLAQVSYY9 pKa = 10.18TGNGSTTQYY18 pKa = 10.87SITFPYY24 pKa = 10.18IDD26 pKa = 3.6VTHH29 pKa = 6.27VKK31 pKa = 10.38AYY33 pKa = 11.1LNGTLTSAYY42 pKa = 8.52TISSSTLTFTTAPANGVVIRR62 pKa = 11.84IEE64 pKa = 4.06RR65 pKa = 11.84EE66 pKa = 3.93TPNDD70 pKa = 3.04NRR72 pKa = 11.84LVDD75 pKa = 4.18FTDD78 pKa = 4.42GSVLTEE84 pKa = 3.73QDD86 pKa = 3.31LDD88 pKa = 3.76RR89 pKa = 11.84SADD92 pKa = 3.46QNFYY96 pKa = 10.27IAQEE100 pKa = 4.05ITDD103 pKa = 4.25DD104 pKa = 3.98SASKK108 pKa = 10.76LGLDD112 pKa = 4.38TDD114 pKa = 4.43DD115 pKa = 5.25KK116 pKa = 11.8YY117 pKa = 11.6NAQSKK122 pKa = 9.09VIKK125 pKa = 10.64NLANPVNDD133 pKa = 3.31TDD135 pKa = 4.1AVNKK139 pKa = 9.93QFISTNLPNINTVAGISANVTTVAGISGNVTTVAGNNANVSTVATNIASVNTVATNIADD198 pKa = 4.21VITVANDD205 pKa = 3.15LNEE208 pKa = 4.92AISEE212 pKa = 4.25IEE214 pKa = 3.99TTANDD219 pKa = 3.39LNEE222 pKa = 4.41AVSEE226 pKa = 4.01IDD228 pKa = 3.48TVGTNIASVQAVGTDD243 pKa = 2.98IANVNTVATDD253 pKa = 3.46IANVNTVATDD263 pKa = 3.46IANVNTVGGAIANVNTVAGANANITTVANANANINTVATDD303 pKa = 3.54IANVNLVGGSIANVNTVATNVANVNTVASNNANINTVASANANITTLAGISADD356 pKa = 2.88ITTVATNSANISTIATDD373 pKa = 3.15IAKK376 pKa = 10.54VITTANDD383 pKa = 3.26LNEE386 pKa = 4.31ATSEE390 pKa = 4.14IEE392 pKa = 4.19VVANAIANVDD402 pKa = 3.29IVGTDD407 pKa = 2.94IADD410 pKa = 3.74VNTVATNIANINAVNSNSSNINAVNSNSANINTVATNDD448 pKa = 3.46ANITTVAGSIANVNTVGSNIGTVNEE473 pKa = 3.84FGEE476 pKa = 4.71RR477 pKa = 11.84YY478 pKa = 9.02RR479 pKa = 11.84VSASAPSTSLDD490 pKa = 3.96LGDD493 pKa = 5.46LYY495 pKa = 11.35FDD497 pKa = 3.82TASNTMKK504 pKa = 10.34VYY506 pKa = 10.95SSGGWINAGSSVNGTADD523 pKa = 3.5RR524 pKa = 11.84YY525 pKa = 10.78KK526 pKa = 9.54YY527 pKa = 8.79TATASQTTFTGADD540 pKa = 3.54DD541 pKa = 3.94NGNTLGYY548 pKa = 10.3DD549 pKa = 3.07AGFLDD554 pKa = 4.67VYY556 pKa = 11.11LNGIRR561 pKa = 11.84LVNGSDD567 pKa = 3.66FTASSGSSIVLTTGASVSDD586 pKa = 3.5ILEE589 pKa = 3.99VVAFGTFQLANFSITDD605 pKa = 3.76ANDD608 pKa = 3.15VPPLGSAGQALVVNSGGTALEE629 pKa = 4.08FANASSAEE637 pKa = 4.19VYY639 pKa = 10.65GFSRR643 pKa = 11.84DD644 pKa = 3.22INGNLIVTTTNQGQDD659 pKa = 3.27SITQSEE665 pKa = 4.84YY666 pKa = 11.53ANFDD670 pKa = 3.59DD671 pKa = 5.21VLFSASGFTFSISNGEE687 pKa = 4.34LIATII692 pKa = 4.25
MM1 pKa = 7.49SFLAQVSYY9 pKa = 10.18TGNGSTTQYY18 pKa = 10.87SITFPYY24 pKa = 10.18IDD26 pKa = 3.6VTHH29 pKa = 6.27VKK31 pKa = 10.38AYY33 pKa = 11.1LNGTLTSAYY42 pKa = 8.52TISSSTLTFTTAPANGVVIRR62 pKa = 11.84IEE64 pKa = 4.06RR65 pKa = 11.84EE66 pKa = 3.93TPNDD70 pKa = 3.04NRR72 pKa = 11.84LVDD75 pKa = 4.18FTDD78 pKa = 4.42GSVLTEE84 pKa = 3.73QDD86 pKa = 3.31LDD88 pKa = 3.76RR89 pKa = 11.84SADD92 pKa = 3.46QNFYY96 pKa = 10.27IAQEE100 pKa = 4.05ITDD103 pKa = 4.25DD104 pKa = 3.98SASKK108 pKa = 10.76LGLDD112 pKa = 4.38TDD114 pKa = 4.43DD115 pKa = 5.25KK116 pKa = 11.8YY117 pKa = 11.6NAQSKK122 pKa = 9.09VIKK125 pKa = 10.64NLANPVNDD133 pKa = 3.31TDD135 pKa = 4.1AVNKK139 pKa = 9.93QFISTNLPNINTVAGISANVTTVAGISGNVTTVAGNNANVSTVATNIASVNTVATNIADD198 pKa = 4.21VITVANDD205 pKa = 3.15LNEE208 pKa = 4.92AISEE212 pKa = 4.25IEE214 pKa = 3.99TTANDD219 pKa = 3.39LNEE222 pKa = 4.41AVSEE226 pKa = 4.01IDD228 pKa = 3.48TVGTNIASVQAVGTDD243 pKa = 2.98IANVNTVATDD253 pKa = 3.46IANVNTVATDD263 pKa = 3.46IANVNTVGGAIANVNTVAGANANITTVANANANINTVATDD303 pKa = 3.54IANVNLVGGSIANVNTVATNVANVNTVASNNANINTVASANANITTLAGISADD356 pKa = 2.88ITTVATNSANISTIATDD373 pKa = 3.15IAKK376 pKa = 10.54VITTANDD383 pKa = 3.26LNEE386 pKa = 4.31ATSEE390 pKa = 4.14IEE392 pKa = 4.19VVANAIANVDD402 pKa = 3.29IVGTDD407 pKa = 2.94IADD410 pKa = 3.74VNTVATNIANINAVNSNSSNINAVNSNSANINTVATNDD448 pKa = 3.46ANITTVAGSIANVNTVGSNIGTVNEE473 pKa = 3.84FGEE476 pKa = 4.71RR477 pKa = 11.84YY478 pKa = 9.02RR479 pKa = 11.84VSASAPSTSLDD490 pKa = 3.96LGDD493 pKa = 5.46LYY495 pKa = 11.35FDD497 pKa = 3.82TASNTMKK504 pKa = 10.34VYY506 pKa = 10.95SSGGWINAGSSVNGTADD523 pKa = 3.5RR524 pKa = 11.84YY525 pKa = 10.78KK526 pKa = 9.54YY527 pKa = 8.79TATASQTTFTGADD540 pKa = 3.54DD541 pKa = 3.94NGNTLGYY548 pKa = 10.3DD549 pKa = 3.07AGFLDD554 pKa = 4.67VYY556 pKa = 11.11LNGIRR561 pKa = 11.84LVNGSDD567 pKa = 3.66FTASSGSSIVLTTGASVSDD586 pKa = 3.5ILEE589 pKa = 3.99VVAFGTFQLANFSITDD605 pKa = 3.76ANDD608 pKa = 3.15VPPLGSAGQALVVNSGGTALEE629 pKa = 4.08FANASSAEE637 pKa = 4.19VYY639 pKa = 10.65GFSRR643 pKa = 11.84DD644 pKa = 3.22INGNLIVTTTNQGQDD659 pKa = 3.27SITQSEE665 pKa = 4.84YY666 pKa = 11.53ANFDD670 pKa = 3.59DD671 pKa = 5.21VLFSASGFTFSISNGEE687 pKa = 4.34LIATII692 pKa = 4.25
Molecular weight: 70.76 kDa
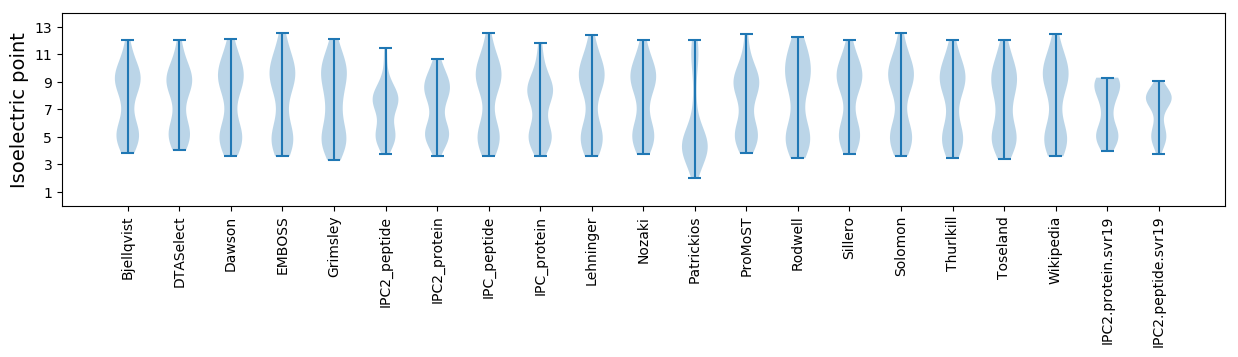
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1NU31|A0A4Y1NU31_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC025P OX=2259657 GN=P025_gp10 PE=4 SV=1
MM1 pKa = 7.8CNFKK5 pKa = 10.91APSPPPAPEE14 pKa = 5.26PIPEE18 pKa = 4.24TPPVVTQATTTQKK31 pKa = 11.16APATARR37 pKa = 11.84NEE39 pKa = 4.19SGVNRR44 pKa = 11.84NMASEE49 pKa = 4.53VSRR52 pKa = 11.84KK53 pKa = 8.8RR54 pKa = 11.84QGRR57 pKa = 11.84GSLRR61 pKa = 11.84IPLASLGSGSGLNFPTAA78 pKa = 3.57
MM1 pKa = 7.8CNFKK5 pKa = 10.91APSPPPAPEE14 pKa = 5.26PIPEE18 pKa = 4.24TPPVVTQATTTQKK31 pKa = 11.16APATARR37 pKa = 11.84NEE39 pKa = 4.19SGVNRR44 pKa = 11.84NMASEE49 pKa = 4.53VSRR52 pKa = 11.84KK53 pKa = 8.8RR54 pKa = 11.84QGRR57 pKa = 11.84GSLRR61 pKa = 11.84IPLASLGSGSGLNFPTAA78 pKa = 3.57
Molecular weight: 8.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11728 |
40 |
1246 |
239.3 |
26.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.855 ± 0.525 | 0.929 ± 0.187 |
5.585 ± 0.251 | 6.344 ± 0.451 |
4.221 ± 0.326 | 6.804 ± 0.779 |
1.228 ± 0.213 | 6.949 ± 0.32 |
8.245 ± 0.76 | 8.058 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.918 ± 0.197 | 6.693 ± 0.48 |
2.993 ± 0.235 | 4.246 ± 0.257 |
4.033 ± 0.266 | 7.171 ± 0.547 |
6.881 ± 0.71 | 6.003 ± 0.355 |
0.946 ± 0.118 | 3.897 ± 0.179 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
