
Brevifollis gellanilyticus
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Verrucomicrobiae; Verrucomicrobiales; Verrucomicrobiaceae; Brevifollis
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
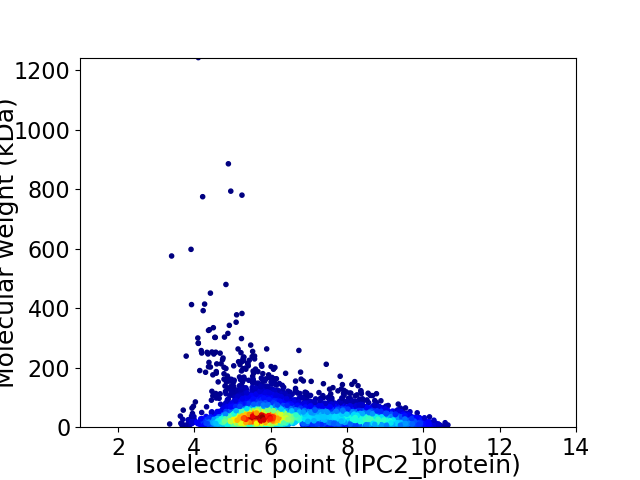
Virtual 2D-PAGE plot for 5783 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A512MDV2|A0A512MDV2_9BACT Non-specific serine/threonine protein kinase OS=Brevifollis gellanilyticus OX=748831 GN=BGE01nite_41630 PE=4 SV=1
MM1 pKa = 7.57IFLIFSGAHH10 pKa = 5.33FAWGYY15 pKa = 10.39LGSFEE20 pKa = 5.54EE21 pKa = 4.23NDD23 pKa = 4.12GYY25 pKa = 10.94RR26 pKa = 11.84VPSSGNILSLNFAGDD41 pKa = 3.39AQFYY45 pKa = 10.53LGNNPTNGLTGIVAPGAYY63 pKa = 9.27PSTLGDD69 pKa = 3.51ATHH72 pKa = 6.72GLDD75 pKa = 3.45VSRR78 pKa = 11.84YY79 pKa = 8.68NAGQYY84 pKa = 10.58GGGGGGPGGNGTDD97 pKa = 3.35IQDD100 pKa = 3.55NTGLWRR106 pKa = 11.84ALSGGRR112 pKa = 11.84LNEE115 pKa = 4.33DD116 pKa = 2.46QGAPYY121 pKa = 10.47YY122 pKa = 10.8LGGPFQRR129 pKa = 11.84DD130 pKa = 3.41YY131 pKa = 11.28IAGYY135 pKa = 10.12NYY137 pKa = 10.39GSARR141 pKa = 11.84TGSQVLNLLAYY152 pKa = 10.11DD153 pKa = 3.55EE154 pKa = 4.34NLRR157 pKa = 11.84YY158 pKa = 9.9SYY160 pKa = 11.45SLDD163 pKa = 3.29SRR165 pKa = 11.84DD166 pKa = 3.4FDD168 pKa = 4.87GINPATTSDD177 pKa = 3.37MRR179 pKa = 11.84IEE181 pKa = 4.1ASFWTCPTDD190 pKa = 3.94ADD192 pKa = 4.01DD193 pKa = 6.66DD194 pKa = 3.9IAIGVNTVGLSFRR207 pKa = 11.84DD208 pKa = 3.4SMDD211 pKa = 3.39QILVDD216 pKa = 4.05IGYY219 pKa = 8.65TGDD222 pKa = 3.7NFLQYY227 pKa = 10.45RR228 pKa = 11.84IGGSSTWQVTALNLGATGWSQINLVLDD255 pKa = 3.92TQANSVSLAARR266 pKa = 11.84AYY268 pKa = 10.83SDD270 pKa = 2.94ITGTLGTDD278 pKa = 3.14TTVFTDD284 pKa = 3.21QLLGFNTDD292 pKa = 2.61NVTQIEE298 pKa = 4.34WTATDD303 pKa = 2.95VDD305 pKa = 3.94GFKK308 pKa = 11.22NFFDD312 pKa = 5.4DD313 pKa = 5.3FDD315 pKa = 4.01FTLMSVVPEE324 pKa = 4.34PGSLTFLVLAWLYY337 pKa = 10.1SMRR340 pKa = 11.84RR341 pKa = 11.84KK342 pKa = 10.11RR343 pKa = 11.84EE344 pKa = 3.82SS345 pKa = 2.92
MM1 pKa = 7.57IFLIFSGAHH10 pKa = 5.33FAWGYY15 pKa = 10.39LGSFEE20 pKa = 5.54EE21 pKa = 4.23NDD23 pKa = 4.12GYY25 pKa = 10.94RR26 pKa = 11.84VPSSGNILSLNFAGDD41 pKa = 3.39AQFYY45 pKa = 10.53LGNNPTNGLTGIVAPGAYY63 pKa = 9.27PSTLGDD69 pKa = 3.51ATHH72 pKa = 6.72GLDD75 pKa = 3.45VSRR78 pKa = 11.84YY79 pKa = 8.68NAGQYY84 pKa = 10.58GGGGGGPGGNGTDD97 pKa = 3.35IQDD100 pKa = 3.55NTGLWRR106 pKa = 11.84ALSGGRR112 pKa = 11.84LNEE115 pKa = 4.33DD116 pKa = 2.46QGAPYY121 pKa = 10.47YY122 pKa = 10.8LGGPFQRR129 pKa = 11.84DD130 pKa = 3.41YY131 pKa = 11.28IAGYY135 pKa = 10.12NYY137 pKa = 10.39GSARR141 pKa = 11.84TGSQVLNLLAYY152 pKa = 10.11DD153 pKa = 3.55EE154 pKa = 4.34NLRR157 pKa = 11.84YY158 pKa = 9.9SYY160 pKa = 11.45SLDD163 pKa = 3.29SRR165 pKa = 11.84DD166 pKa = 3.4FDD168 pKa = 4.87GINPATTSDD177 pKa = 3.37MRR179 pKa = 11.84IEE181 pKa = 4.1ASFWTCPTDD190 pKa = 3.94ADD192 pKa = 4.01DD193 pKa = 6.66DD194 pKa = 3.9IAIGVNTVGLSFRR207 pKa = 11.84DD208 pKa = 3.4SMDD211 pKa = 3.39QILVDD216 pKa = 4.05IGYY219 pKa = 8.65TGDD222 pKa = 3.7NFLQYY227 pKa = 10.45RR228 pKa = 11.84IGGSSTWQVTALNLGATGWSQINLVLDD255 pKa = 3.92TQANSVSLAARR266 pKa = 11.84AYY268 pKa = 10.83SDD270 pKa = 2.94ITGTLGTDD278 pKa = 3.14TTVFTDD284 pKa = 3.21QLLGFNTDD292 pKa = 2.61NVTQIEE298 pKa = 4.34WTATDD303 pKa = 2.95VDD305 pKa = 3.94GFKK308 pKa = 11.22NFFDD312 pKa = 5.4DD313 pKa = 5.3FDD315 pKa = 4.01FTLMSVVPEE324 pKa = 4.34PGSLTFLVLAWLYY337 pKa = 10.1SMRR340 pKa = 11.84RR341 pKa = 11.84KK342 pKa = 10.11RR343 pKa = 11.84EE344 pKa = 3.82SS345 pKa = 2.92
Molecular weight: 37.29 kDa
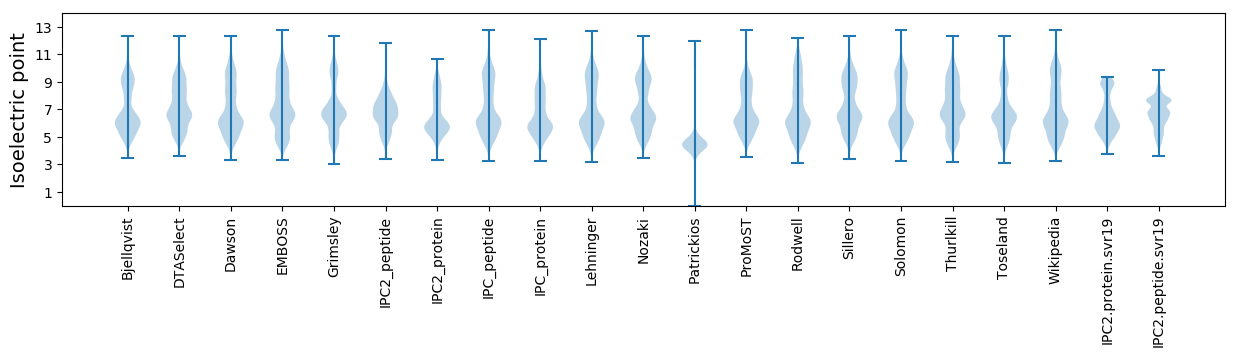
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A512MIS1|A0A512MIS1_9BACT Uncharacterized protein OS=Brevifollis gellanilyticus OX=748831 GN=BGE01nite_54920 PE=4 SV=1
MM1 pKa = 7.1VAGAQQVRR9 pKa = 11.84RR10 pKa = 11.84ARR12 pKa = 11.84EE13 pKa = 3.62AAGLRR18 pKa = 11.84THH20 pKa = 7.14CAAKK24 pKa = 9.24STLPQLLQAMIQQALAGSGSLGHH47 pKa = 6.69HH48 pKa = 6.57LRR50 pKa = 11.84LVMNTRR56 pKa = 11.84ISDD59 pKa = 3.45SAAIQRR65 pKa = 11.84RR66 pKa = 11.84ASLNWEE72 pKa = 4.97FYY74 pKa = 9.96QEE76 pKa = 3.94LFGRR80 pKa = 11.84VLKK83 pKa = 10.5PLARR87 pKa = 11.84PKK89 pKa = 10.58DD90 pKa = 3.64HH91 pKa = 7.4AEE93 pKa = 3.94SFHH96 pKa = 7.28AGLRR100 pKa = 11.84LLALDD105 pKa = 4.77GSHH108 pKa = 7.52FSLRR112 pKa = 11.84NSAAVMAMTRR122 pKa = 11.84PRR124 pKa = 11.84PGNQRR129 pKa = 11.84ASAGAFMKK137 pKa = 10.31WSTAVLLEE145 pKa = 4.77LGTHH149 pKa = 6.62APLSVACSTLGMEE162 pKa = 4.41RR163 pKa = 11.84MEE165 pKa = 4.25MEE167 pKa = 3.39IDD169 pKa = 2.85IARR172 pKa = 11.84RR173 pKa = 11.84TLPTLATLGPSLLLADD189 pKa = 4.7RR190 pKa = 11.84LYY192 pKa = 11.29GCASFMLEE200 pKa = 4.85VMEE203 pKa = 4.14QGQGHH208 pKa = 6.33AHH210 pKa = 6.42CLMRR214 pKa = 11.84VRR216 pKa = 11.84DD217 pKa = 4.03NLRR220 pKa = 11.84AQQLEE225 pKa = 4.21VLHH228 pKa = 6.96DD229 pKa = 4.31GSALMRR235 pKa = 11.84VRR237 pKa = 11.84SDD239 pKa = 2.85TKK241 pKa = 10.99ARR243 pKa = 11.84AKK245 pKa = 10.29AASCEE250 pKa = 4.09MVVRR254 pKa = 11.84EE255 pKa = 4.13LRR257 pKa = 11.84AEE259 pKa = 3.97VRR261 pKa = 11.84RR262 pKa = 11.84TGGKK266 pKa = 9.59AQTLRR271 pKa = 11.84LWTTLLDD278 pKa = 4.0ARR280 pKa = 11.84KK281 pKa = 9.38HH282 pKa = 4.98PAEE285 pKa = 4.44EE286 pKa = 3.97LLKK289 pKa = 10.79LYY291 pKa = 9.91AQRR294 pKa = 11.84WEE296 pKa = 3.83QEE298 pKa = 3.86LFFRR302 pKa = 11.84EE303 pKa = 4.55LKK305 pKa = 10.12RR306 pKa = 11.84HH307 pKa = 5.34TGATSMLQADD317 pKa = 4.55SEE319 pKa = 4.47QTAQGAFAALVLAASEE335 pKa = 4.32VAQRR339 pKa = 11.84RR340 pKa = 11.84VEE342 pKa = 4.05AATEE346 pKa = 3.72ADD348 pKa = 4.08LPVLRR353 pKa = 11.84LSIAKK358 pKa = 8.51MGQMLGQLTSMLQSAGDD375 pKa = 3.82VLTPAQRR382 pKa = 11.84AAMSHH387 pKa = 5.79KK388 pKa = 8.64WLQVMVRR395 pKa = 11.84EE396 pKa = 4.18AVIPPRR402 pKa = 11.84RR403 pKa = 11.84ARR405 pKa = 11.84SCQHH409 pKa = 5.54GLRR412 pKa = 11.84RR413 pKa = 11.84PHH415 pKa = 6.64SPWPVIRR422 pKa = 11.84KK423 pKa = 8.89RR424 pKa = 11.84VFIDD428 pKa = 3.47PEE430 pKa = 4.14VLLTLLPATSPP441 pKa = 3.52
MM1 pKa = 7.1VAGAQQVRR9 pKa = 11.84RR10 pKa = 11.84ARR12 pKa = 11.84EE13 pKa = 3.62AAGLRR18 pKa = 11.84THH20 pKa = 7.14CAAKK24 pKa = 9.24STLPQLLQAMIQQALAGSGSLGHH47 pKa = 6.69HH48 pKa = 6.57LRR50 pKa = 11.84LVMNTRR56 pKa = 11.84ISDD59 pKa = 3.45SAAIQRR65 pKa = 11.84RR66 pKa = 11.84ASLNWEE72 pKa = 4.97FYY74 pKa = 9.96QEE76 pKa = 3.94LFGRR80 pKa = 11.84VLKK83 pKa = 10.5PLARR87 pKa = 11.84PKK89 pKa = 10.58DD90 pKa = 3.64HH91 pKa = 7.4AEE93 pKa = 3.94SFHH96 pKa = 7.28AGLRR100 pKa = 11.84LLALDD105 pKa = 4.77GSHH108 pKa = 7.52FSLRR112 pKa = 11.84NSAAVMAMTRR122 pKa = 11.84PRR124 pKa = 11.84PGNQRR129 pKa = 11.84ASAGAFMKK137 pKa = 10.31WSTAVLLEE145 pKa = 4.77LGTHH149 pKa = 6.62APLSVACSTLGMEE162 pKa = 4.41RR163 pKa = 11.84MEE165 pKa = 4.25MEE167 pKa = 3.39IDD169 pKa = 2.85IARR172 pKa = 11.84RR173 pKa = 11.84TLPTLATLGPSLLLADD189 pKa = 4.7RR190 pKa = 11.84LYY192 pKa = 11.29GCASFMLEE200 pKa = 4.85VMEE203 pKa = 4.14QGQGHH208 pKa = 6.33AHH210 pKa = 6.42CLMRR214 pKa = 11.84VRR216 pKa = 11.84DD217 pKa = 4.03NLRR220 pKa = 11.84AQQLEE225 pKa = 4.21VLHH228 pKa = 6.96DD229 pKa = 4.31GSALMRR235 pKa = 11.84VRR237 pKa = 11.84SDD239 pKa = 2.85TKK241 pKa = 10.99ARR243 pKa = 11.84AKK245 pKa = 10.29AASCEE250 pKa = 4.09MVVRR254 pKa = 11.84EE255 pKa = 4.13LRR257 pKa = 11.84AEE259 pKa = 3.97VRR261 pKa = 11.84RR262 pKa = 11.84TGGKK266 pKa = 9.59AQTLRR271 pKa = 11.84LWTTLLDD278 pKa = 4.0ARR280 pKa = 11.84KK281 pKa = 9.38HH282 pKa = 4.98PAEE285 pKa = 4.44EE286 pKa = 3.97LLKK289 pKa = 10.79LYY291 pKa = 9.91AQRR294 pKa = 11.84WEE296 pKa = 3.83QEE298 pKa = 3.86LFFRR302 pKa = 11.84EE303 pKa = 4.55LKK305 pKa = 10.12RR306 pKa = 11.84HH307 pKa = 5.34TGATSMLQADD317 pKa = 4.55SEE319 pKa = 4.47QTAQGAFAALVLAASEE335 pKa = 4.32VAQRR339 pKa = 11.84RR340 pKa = 11.84VEE342 pKa = 4.05AATEE346 pKa = 3.72ADD348 pKa = 4.08LPVLRR353 pKa = 11.84LSIAKK358 pKa = 8.51MGQMLGQLTSMLQSAGDD375 pKa = 3.82VLTPAQRR382 pKa = 11.84AAMSHH387 pKa = 5.79KK388 pKa = 8.64WLQVMVRR395 pKa = 11.84EE396 pKa = 4.18AVIPPRR402 pKa = 11.84RR403 pKa = 11.84ARR405 pKa = 11.84SCQHH409 pKa = 5.54GLRR412 pKa = 11.84RR413 pKa = 11.84PHH415 pKa = 6.64SPWPVIRR422 pKa = 11.84KK423 pKa = 8.89RR424 pKa = 11.84VFIDD428 pKa = 3.47PEE430 pKa = 4.14VLLTLLPATSPP441 pKa = 3.52
Molecular weight: 48.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2267602 |
39 |
12113 |
392.1 |
42.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.309 ± 0.038 | 0.927 ± 0.014 |
5.427 ± 0.031 | 5.722 ± 0.055 |
3.77 ± 0.018 | 8.326 ± 0.069 |
2.296 ± 0.027 | 4.758 ± 0.023 |
4.6 ± 0.056 | 9.865 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.346 ± 0.022 | 3.331 ± 0.058 |
5.471 ± 0.031 | 3.62 ± 0.025 |
5.717 ± 0.042 | 6.243 ± 0.042 |
6.343 ± 0.094 | 6.94 ± 0.029 |
1.602 ± 0.019 | 2.387 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
