
Sphingopyxis alaskensis (strain DSM 13593 / LMG 18877 / RB2256) (Sphingomonas alaskensis)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingopyxis; Sphingopyxis alaskensis
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
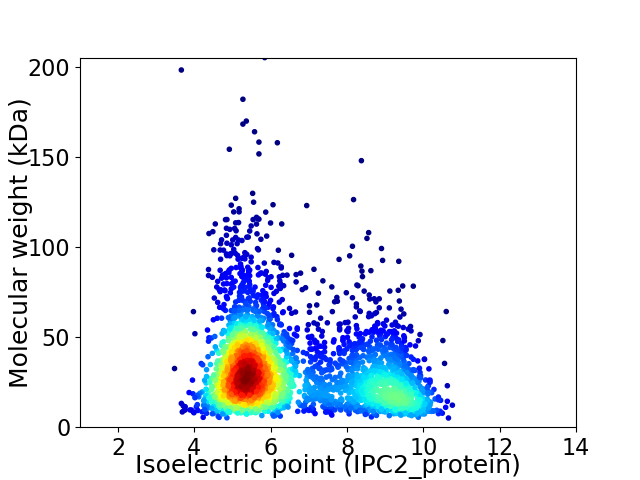
Virtual 2D-PAGE plot for 3184 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1GQW0|Q1GQW0_SPHAL Ferrochelatase OS=Sphingopyxis alaskensis (strain DSM 13593 / LMG 18877 / RB2256) OX=317655 GN=hemH PE=3 SV=1
MM1 pKa = 7.0QSVRR5 pKa = 11.84ACIMLVWLALTFASNPVTAQTTTSYY30 pKa = 10.02STTTTGTISEE40 pKa = 4.55SATPCSAPMLRR51 pKa = 11.84SFTVAANAQVSDD63 pKa = 3.95VNIGVQFAHH72 pKa = 7.09TYY74 pKa = 10.21RR75 pKa = 11.84GDD77 pKa = 3.07IRR79 pKa = 11.84ATLTSPAGTVVDD91 pKa = 5.93LITNVGASADD101 pKa = 3.81NLNVLFDD108 pKa = 4.82DD109 pKa = 5.24GAAASITTHH118 pKa = 6.27NANDD122 pKa = 3.77STATAPPYY130 pKa = 10.48QRR132 pKa = 11.84TFAPQGSLAAFNGQASGGTWQLSICDD158 pKa = 4.45SLNGDD163 pKa = 3.33AGTFTRR169 pKa = 11.84ADD171 pKa = 3.55LTLTTVPLSPYY182 pKa = 10.78ADD184 pKa = 3.59LSLAKK189 pKa = 8.98TVSNAAPASGAIINYY204 pKa = 8.45VLSVTNAGASTLTATGVTVQDD225 pKa = 3.59TLPAGFAFTGASGFGSYY242 pKa = 10.96NSTTGVWTVGSVPPGVTRR260 pKa = 11.84TLTISGTVAASAGASVTNSAEE281 pKa = 3.92IAASSAFDD289 pKa = 4.13PDD291 pKa = 3.72STPGNGAAGEE301 pKa = 4.32DD302 pKa = 4.36DD303 pKa = 4.18GDD305 pKa = 4.04SASFTVSGTRR315 pKa = 11.84VAGTPPTLVCPVGTTALDD333 pKa = 3.36WDD335 pKa = 4.71TIAWNAGTTSASPPLTAIGTASIGIAVSGGVFLSNATYY373 pKa = 10.51GGQSPTRR380 pKa = 11.84QNIVTGGLSPAQYY393 pKa = 10.79SIFQLVDD400 pKa = 3.78FTSQAGSVATTISLPTAVPGAQFRR424 pKa = 11.84LFDD427 pKa = 3.33VDD429 pKa = 3.59YY430 pKa = 11.14AAGQFADD437 pKa = 3.61RR438 pKa = 11.84VTVTGSYY445 pKa = 10.45NGAPVTPILTNGTANYY461 pKa = 10.26VIGNSAYY468 pKa = 10.55GDD470 pKa = 3.84VLSTDD475 pKa = 3.08ASGNGNITVTFAAPVDD491 pKa = 4.25TIVIDD496 pKa = 4.04YY497 pKa = 9.9GSHH500 pKa = 6.53ALAPANPGQQGMALHH515 pKa = 7.36DD516 pKa = 4.74IIFCRR521 pKa = 11.84PTANLTVAKK530 pKa = 9.05TSSIVSDD537 pKa = 3.73PVGGATNPKK546 pKa = 9.23AIPGATMRR554 pKa = 11.84YY555 pKa = 9.18CILVTNNGSGTATAINIADD574 pKa = 4.4ALPARR579 pKa = 11.84TTFVPGSLRR588 pKa = 11.84SGTSCAAATTVEE600 pKa = 4.72DD601 pKa = 4.78DD602 pKa = 3.68NASGGDD608 pKa = 3.38EE609 pKa = 4.31SDD611 pKa = 3.53PFGASFAGSAIAASTATLAPGGAMAIAFTVTIDD644 pKa = 3.0
MM1 pKa = 7.0QSVRR5 pKa = 11.84ACIMLVWLALTFASNPVTAQTTTSYY30 pKa = 10.02STTTTGTISEE40 pKa = 4.55SATPCSAPMLRR51 pKa = 11.84SFTVAANAQVSDD63 pKa = 3.95VNIGVQFAHH72 pKa = 7.09TYY74 pKa = 10.21RR75 pKa = 11.84GDD77 pKa = 3.07IRR79 pKa = 11.84ATLTSPAGTVVDD91 pKa = 5.93LITNVGASADD101 pKa = 3.81NLNVLFDD108 pKa = 4.82DD109 pKa = 5.24GAAASITTHH118 pKa = 6.27NANDD122 pKa = 3.77STATAPPYY130 pKa = 10.48QRR132 pKa = 11.84TFAPQGSLAAFNGQASGGTWQLSICDD158 pKa = 4.45SLNGDD163 pKa = 3.33AGTFTRR169 pKa = 11.84ADD171 pKa = 3.55LTLTTVPLSPYY182 pKa = 10.78ADD184 pKa = 3.59LSLAKK189 pKa = 8.98TVSNAAPASGAIINYY204 pKa = 8.45VLSVTNAGASTLTATGVTVQDD225 pKa = 3.59TLPAGFAFTGASGFGSYY242 pKa = 10.96NSTTGVWTVGSVPPGVTRR260 pKa = 11.84TLTISGTVAASAGASVTNSAEE281 pKa = 3.92IAASSAFDD289 pKa = 4.13PDD291 pKa = 3.72STPGNGAAGEE301 pKa = 4.32DD302 pKa = 4.36DD303 pKa = 4.18GDD305 pKa = 4.04SASFTVSGTRR315 pKa = 11.84VAGTPPTLVCPVGTTALDD333 pKa = 3.36WDD335 pKa = 4.71TIAWNAGTTSASPPLTAIGTASIGIAVSGGVFLSNATYY373 pKa = 10.51GGQSPTRR380 pKa = 11.84QNIVTGGLSPAQYY393 pKa = 10.79SIFQLVDD400 pKa = 3.78FTSQAGSVATTISLPTAVPGAQFRR424 pKa = 11.84LFDD427 pKa = 3.33VDD429 pKa = 3.59YY430 pKa = 11.14AAGQFADD437 pKa = 3.61RR438 pKa = 11.84VTVTGSYY445 pKa = 10.45NGAPVTPILTNGTANYY461 pKa = 10.26VIGNSAYY468 pKa = 10.55GDD470 pKa = 3.84VLSTDD475 pKa = 3.08ASGNGNITVTFAAPVDD491 pKa = 4.25TIVIDD496 pKa = 4.04YY497 pKa = 9.9GSHH500 pKa = 6.53ALAPANPGQQGMALHH515 pKa = 7.36DD516 pKa = 4.74IIFCRR521 pKa = 11.84PTANLTVAKK530 pKa = 9.05TSSIVSDD537 pKa = 3.73PVGGATNPKK546 pKa = 9.23AIPGATMRR554 pKa = 11.84YY555 pKa = 9.18CILVTNNGSGTATAINIADD574 pKa = 4.4ALPARR579 pKa = 11.84TTFVPGSLRR588 pKa = 11.84SGTSCAAATTVEE600 pKa = 4.72DD601 pKa = 4.78DD602 pKa = 3.68NASGGDD608 pKa = 3.38EE609 pKa = 4.31SDD611 pKa = 3.53PFGASFAGSAIAASTATLAPGGAMAIAFTVTIDD644 pKa = 3.0
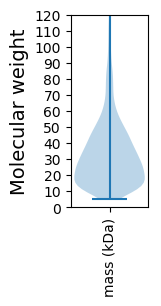
Molecular weight: 64.09 kDa
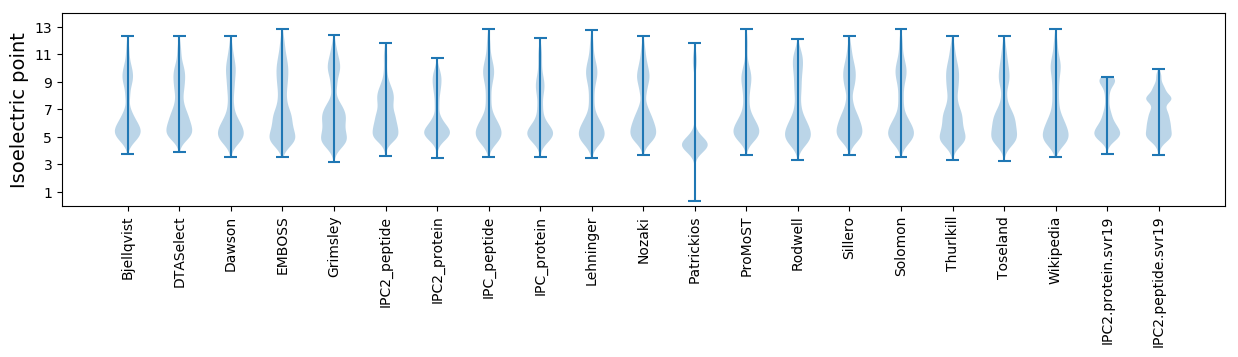
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1GQ97|Q1GQ97_SPHAL Oxidoreductase FAD-binding region OS=Sphingopyxis alaskensis (strain DSM 13593 / LMG 18877 / RB2256) OX=317655 GN=Sala_2468 PE=4 SV=1
MM1 pKa = 7.52SSPRR5 pKa = 11.84GSDD8 pKa = 3.35PASGAGDD15 pKa = 3.17WSGNWRR21 pKa = 11.84TLVGLWAIPGLAMLAAMWLEE41 pKa = 3.91PTPRR45 pKa = 11.84AVIWTVMLASMGAACIMNARR65 pKa = 11.84RR66 pKa = 11.84CGRR69 pKa = 11.84THH71 pKa = 6.99CRR73 pKa = 11.84FTGPFLIGMAILVVAYY89 pKa = 9.92AIGMLPLGPHH99 pKa = 5.86GWGILGGVTLGGFAALWWGSEE120 pKa = 4.11RR121 pKa = 11.84AWGKK125 pKa = 9.76FSRR128 pKa = 11.84SKK130 pKa = 11.18DD131 pKa = 3.26KK132 pKa = 11.16GVSRR136 pKa = 11.84SCC138 pKa = 3.95
MM1 pKa = 7.52SSPRR5 pKa = 11.84GSDD8 pKa = 3.35PASGAGDD15 pKa = 3.17WSGNWRR21 pKa = 11.84TLVGLWAIPGLAMLAAMWLEE41 pKa = 3.91PTPRR45 pKa = 11.84AVIWTVMLASMGAACIMNARR65 pKa = 11.84RR66 pKa = 11.84CGRR69 pKa = 11.84THH71 pKa = 6.99CRR73 pKa = 11.84FTGPFLIGMAILVVAYY89 pKa = 9.92AIGMLPLGPHH99 pKa = 5.86GWGILGGVTLGGFAALWWGSEE120 pKa = 4.11RR121 pKa = 11.84AWGKK125 pKa = 9.76FSRR128 pKa = 11.84SKK130 pKa = 11.18DD131 pKa = 3.26KK132 pKa = 11.16GVSRR136 pKa = 11.84SCC138 pKa = 3.95
Molecular weight: 14.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1011156 |
41 |
2090 |
317.6 |
34.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.039 ± 0.071 | 0.805 ± 0.015 |
6.197 ± 0.035 | 5.336 ± 0.04 |
3.529 ± 0.026 | 8.921 ± 0.045 |
2.015 ± 0.02 | 5.113 ± 0.031 |
2.912 ± 0.036 | 9.765 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.484 ± 0.019 | 2.391 ± 0.025 |
5.383 ± 0.035 | 2.877 ± 0.021 |
7.649 ± 0.046 | 4.969 ± 0.031 |
5.021 ± 0.027 | 7.021 ± 0.035 |
1.462 ± 0.018 | 2.111 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
