
Apis mellifera associated microvirus 58
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
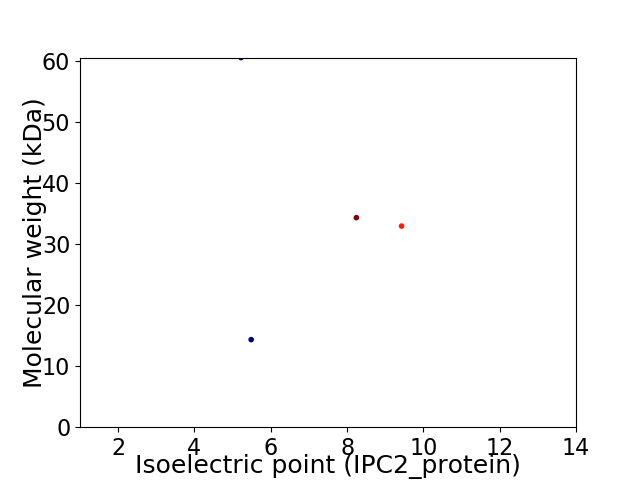
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTT7|A0A3S8UTT7_9VIRU Uncharacterized protein OS=Apis mellifera associated microvirus 58 OX=2494789 PE=4 SV=1
MM1 pKa = 7.38NLFNSVKK8 pKa = 7.63MTRR11 pKa = 11.84PKK13 pKa = 11.2GNTFDD18 pKa = 4.97LSHH21 pKa = 7.21DD22 pKa = 4.07LKK24 pKa = 11.23FSLDD28 pKa = 3.54AGKK31 pKa = 10.39LIPIINQAILPGDD44 pKa = 3.62SWNLSAAQMIRR55 pKa = 11.84VAPMIAPIMHH65 pKa = 6.46RR66 pKa = 11.84VDD68 pKa = 3.38VYY70 pKa = 8.41THH72 pKa = 6.56FFFVPNRR79 pKa = 11.84LLWKK83 pKa = 9.66GWQEE87 pKa = 4.59FITGGNGNDD96 pKa = 3.95PAPVHH101 pKa = 6.56PYY103 pKa = 10.45VILNNKK109 pKa = 5.73QTTVGKK115 pKa = 8.8LTDD118 pKa = 3.62YY119 pKa = 11.19LGIPTVPPASIPVPNPNTKK138 pKa = 9.88VSALDD143 pKa = 3.32IAAVGLVRR151 pKa = 11.84NEE153 pKa = 4.08YY154 pKa = 11.47YY155 pKa = 9.79MDD157 pKa = 4.01QNLQTPWDD165 pKa = 3.81VEE167 pKa = 4.54LNDD170 pKa = 4.64GDD172 pKa = 4.11NTSLLVAAGLIADD185 pKa = 4.4GTTGVGGDD193 pKa = 3.37PFRR196 pKa = 11.84RR197 pKa = 11.84SWEE200 pKa = 3.76HH201 pKa = 7.53DD202 pKa = 3.77YY203 pKa = 9.5FTAALPEE210 pKa = 4.29AQKK213 pKa = 11.22GPAATLPLGSEE224 pKa = 4.01ADD226 pKa = 3.84IIFDD230 pKa = 4.55PNGDD234 pKa = 3.63PSKK237 pKa = 11.29VFDD240 pKa = 3.72VSGNPLSINGHH251 pKa = 5.53LGKK254 pKa = 10.5SGSNDD259 pKa = 4.41LITEE263 pKa = 4.36DD264 pKa = 4.19ALNPGTFVDD273 pKa = 3.99TTLDD277 pKa = 3.1NSAQLKK283 pKa = 10.47ADD285 pKa = 4.3LSTATAATINVVRR298 pKa = 11.84LAFQLQRR305 pKa = 11.84WLEE308 pKa = 4.06RR309 pKa = 11.84NNIAGSRR316 pKa = 11.84YY317 pKa = 9.17IEE319 pKa = 4.4TILAHH324 pKa = 6.14FGVRR328 pKa = 11.84SSDD331 pKa = 3.19KK332 pKa = 10.06TLQRR336 pKa = 11.84PQYY339 pKa = 9.98LGGGKK344 pKa = 9.49SPVMISEE351 pKa = 4.27VLQTSEE357 pKa = 5.34SVTTPQGNLAGHH369 pKa = 6.93GLNVGQSHH377 pKa = 6.67NFTTFFEE384 pKa = 4.29EE385 pKa = 4.31HH386 pKa = 6.86GILLGFISVMPKK398 pKa = 9.73PAYY401 pKa = 9.52QQGLPRR407 pKa = 11.84NMFKK411 pKa = 10.43FDD413 pKa = 4.04RR414 pKa = 11.84IEE416 pKa = 4.19YY417 pKa = 7.9YY418 pKa = 9.65WPSFDD423 pKa = 5.77HH424 pKa = 7.79IGEE427 pKa = 3.99QAIKK431 pKa = 10.38GRR433 pKa = 11.84EE434 pKa = 3.68IFLTGDD440 pKa = 2.95QTYY443 pKa = 11.17DD444 pKa = 3.56DD445 pKa = 5.22KK446 pKa = 11.79DD447 pKa = 2.76WGYY450 pKa = 9.93IPRR453 pKa = 11.84YY454 pKa = 9.12SEE456 pKa = 3.48YY457 pKa = 10.21RR458 pKa = 11.84YY459 pKa = 9.98IPSRR463 pKa = 11.84VAGDD467 pKa = 3.58YY468 pKa = 11.17KK469 pKa = 10.29EE470 pKa = 4.38TLSYY474 pKa = 8.58WHH476 pKa = 7.49WGRR479 pKa = 11.84IFSNEE484 pKa = 3.73PEE486 pKa = 4.22LNEE489 pKa = 4.49EE490 pKa = 5.64FILCDD495 pKa = 3.19PTKK498 pKa = 10.7RR499 pKa = 11.84PFAVQDD505 pKa = 4.43LPDD508 pKa = 4.75SDD510 pKa = 5.61DD511 pKa = 3.99PNTPVRR517 pKa = 11.84DD518 pKa = 3.61WNTMYY523 pKa = 10.68CHH525 pKa = 6.86VVFQIKK531 pKa = 8.57ATRR534 pKa = 11.84RR535 pKa = 11.84MSKK538 pKa = 10.01YY539 pKa = 8.51ATPII543 pKa = 3.38
MM1 pKa = 7.38NLFNSVKK8 pKa = 7.63MTRR11 pKa = 11.84PKK13 pKa = 11.2GNTFDD18 pKa = 4.97LSHH21 pKa = 7.21DD22 pKa = 4.07LKK24 pKa = 11.23FSLDD28 pKa = 3.54AGKK31 pKa = 10.39LIPIINQAILPGDD44 pKa = 3.62SWNLSAAQMIRR55 pKa = 11.84VAPMIAPIMHH65 pKa = 6.46RR66 pKa = 11.84VDD68 pKa = 3.38VYY70 pKa = 8.41THH72 pKa = 6.56FFFVPNRR79 pKa = 11.84LLWKK83 pKa = 9.66GWQEE87 pKa = 4.59FITGGNGNDD96 pKa = 3.95PAPVHH101 pKa = 6.56PYY103 pKa = 10.45VILNNKK109 pKa = 5.73QTTVGKK115 pKa = 8.8LTDD118 pKa = 3.62YY119 pKa = 11.19LGIPTVPPASIPVPNPNTKK138 pKa = 9.88VSALDD143 pKa = 3.32IAAVGLVRR151 pKa = 11.84NEE153 pKa = 4.08YY154 pKa = 11.47YY155 pKa = 9.79MDD157 pKa = 4.01QNLQTPWDD165 pKa = 3.81VEE167 pKa = 4.54LNDD170 pKa = 4.64GDD172 pKa = 4.11NTSLLVAAGLIADD185 pKa = 4.4GTTGVGGDD193 pKa = 3.37PFRR196 pKa = 11.84RR197 pKa = 11.84SWEE200 pKa = 3.76HH201 pKa = 7.53DD202 pKa = 3.77YY203 pKa = 9.5FTAALPEE210 pKa = 4.29AQKK213 pKa = 11.22GPAATLPLGSEE224 pKa = 4.01ADD226 pKa = 3.84IIFDD230 pKa = 4.55PNGDD234 pKa = 3.63PSKK237 pKa = 11.29VFDD240 pKa = 3.72VSGNPLSINGHH251 pKa = 5.53LGKK254 pKa = 10.5SGSNDD259 pKa = 4.41LITEE263 pKa = 4.36DD264 pKa = 4.19ALNPGTFVDD273 pKa = 3.99TTLDD277 pKa = 3.1NSAQLKK283 pKa = 10.47ADD285 pKa = 4.3LSTATAATINVVRR298 pKa = 11.84LAFQLQRR305 pKa = 11.84WLEE308 pKa = 4.06RR309 pKa = 11.84NNIAGSRR316 pKa = 11.84YY317 pKa = 9.17IEE319 pKa = 4.4TILAHH324 pKa = 6.14FGVRR328 pKa = 11.84SSDD331 pKa = 3.19KK332 pKa = 10.06TLQRR336 pKa = 11.84PQYY339 pKa = 9.98LGGGKK344 pKa = 9.49SPVMISEE351 pKa = 4.27VLQTSEE357 pKa = 5.34SVTTPQGNLAGHH369 pKa = 6.93GLNVGQSHH377 pKa = 6.67NFTTFFEE384 pKa = 4.29EE385 pKa = 4.31HH386 pKa = 6.86GILLGFISVMPKK398 pKa = 9.73PAYY401 pKa = 9.52QQGLPRR407 pKa = 11.84NMFKK411 pKa = 10.43FDD413 pKa = 4.04RR414 pKa = 11.84IEE416 pKa = 4.19YY417 pKa = 7.9YY418 pKa = 9.65WPSFDD423 pKa = 5.77HH424 pKa = 7.79IGEE427 pKa = 3.99QAIKK431 pKa = 10.38GRR433 pKa = 11.84EE434 pKa = 3.68IFLTGDD440 pKa = 2.95QTYY443 pKa = 11.17DD444 pKa = 3.56DD445 pKa = 5.22KK446 pKa = 11.79DD447 pKa = 2.76WGYY450 pKa = 9.93IPRR453 pKa = 11.84YY454 pKa = 9.12SEE456 pKa = 3.48YY457 pKa = 10.21RR458 pKa = 11.84YY459 pKa = 9.98IPSRR463 pKa = 11.84VAGDD467 pKa = 3.58YY468 pKa = 11.17KK469 pKa = 10.29EE470 pKa = 4.38TLSYY474 pKa = 8.58WHH476 pKa = 7.49WGRR479 pKa = 11.84IFSNEE484 pKa = 3.73PEE486 pKa = 4.22LNEE489 pKa = 4.49EE490 pKa = 5.64FILCDD495 pKa = 3.19PTKK498 pKa = 10.7RR499 pKa = 11.84PFAVQDD505 pKa = 4.43LPDD508 pKa = 4.75SDD510 pKa = 5.61DD511 pKa = 3.99PNTPVRR517 pKa = 11.84DD518 pKa = 3.61WNTMYY523 pKa = 10.68CHH525 pKa = 6.86VVFQIKK531 pKa = 8.57ATRR534 pKa = 11.84RR535 pKa = 11.84MSKK538 pKa = 10.01YY539 pKa = 8.51ATPII543 pKa = 3.38
Molecular weight: 60.51 kDa
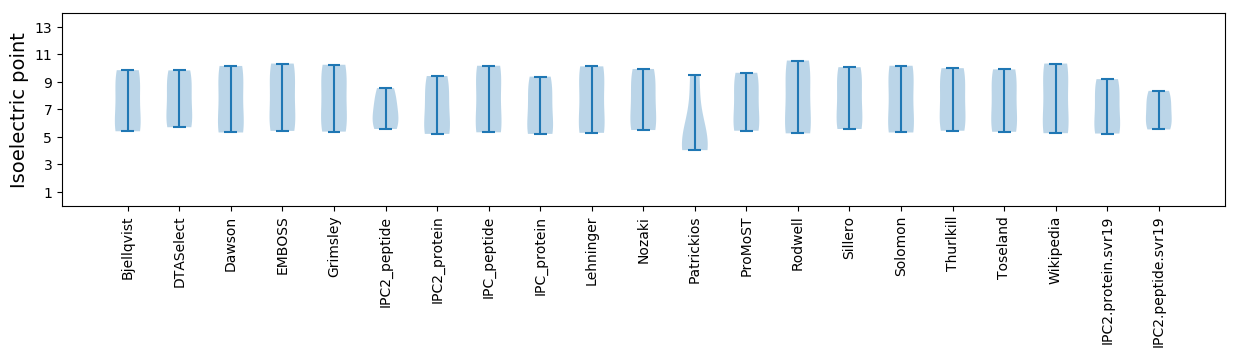
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTY7|A0A3S8UTY7_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 58 OX=2494789 PE=4 SV=1
MM1 pKa = 7.49CFEE4 pKa = 5.61PYY6 pKa = 9.17PLKK9 pKa = 11.04YY10 pKa = 10.22KK11 pKa = 10.22GAKK14 pKa = 9.28LHH16 pKa = 5.76LQGMTIPVPCGKK28 pKa = 10.13CPKK31 pKa = 8.62CTKK34 pKa = 10.46ARR36 pKa = 11.84INSWLFRR43 pKa = 11.84LTIHH47 pKa = 6.2ARR49 pKa = 11.84KK50 pKa = 9.52YY51 pKa = 9.66PYY53 pKa = 10.07PLFITLTYY61 pKa = 10.4DD62 pKa = 3.67DD63 pKa = 4.06KK64 pKa = 11.37HH65 pKa = 7.96LPVHH69 pKa = 6.3SKK71 pKa = 10.04SGRR74 pKa = 11.84VSLDD78 pKa = 2.85KK79 pKa = 10.99SDD81 pKa = 3.45PQKK84 pKa = 9.95WFKK87 pKa = 10.71RR88 pKa = 11.84LRR90 pKa = 11.84KK91 pKa = 9.97SLDD94 pKa = 3.15GQMDD98 pKa = 4.25LSYY101 pKa = 10.91FLVGEE106 pKa = 4.29YY107 pKa = 8.44GTKK110 pKa = 9.94RR111 pKa = 11.84KK112 pKa = 9.7RR113 pKa = 11.84PHH115 pKa = 5.26YY116 pKa = 10.13HH117 pKa = 4.97VLLYY121 pKa = 10.4GLPHH125 pKa = 6.45NKK127 pKa = 9.68SEE129 pKa = 5.46LIYY132 pKa = 9.24KK133 pKa = 8.33TWGLGFTMTLPVLDD147 pKa = 3.58GGMRR151 pKa = 11.84YY152 pKa = 8.56VLKK155 pKa = 11.14YY156 pKa = 8.09MAKK159 pKa = 9.53PRR161 pKa = 11.84SNYY164 pKa = 7.83TSKK167 pKa = 11.02LPEE170 pKa = 4.0FSLMSKK176 pKa = 10.71NIGANYY182 pKa = 7.71LTPQMIRR189 pKa = 11.84YY190 pKa = 8.67HH191 pKa = 8.25LEE193 pKa = 4.26DD194 pKa = 4.27PAHH197 pKa = 6.06TFASLGNGIKK207 pKa = 10.43LALPKK212 pKa = 10.33YY213 pKa = 8.45YY214 pKa = 9.97KK215 pKa = 10.46EE216 pKa = 4.44KK217 pKa = 10.63IFGIDD222 pKa = 2.86TPEE225 pKa = 4.01RR226 pKa = 11.84KK227 pKa = 9.3RR228 pKa = 11.84VTDD231 pKa = 3.58YY232 pKa = 10.61LQKK235 pKa = 10.09RR236 pKa = 11.84AEE238 pKa = 4.15MAQNSYY244 pKa = 10.4LRR246 pKa = 11.84RR247 pKa = 11.84LSLNNRR253 pKa = 11.84CLSQEE258 pKa = 4.25EE259 pKa = 4.37IIKK262 pKa = 10.49LAFLRR267 pKa = 11.84QHH269 pKa = 5.98SLSYY273 pKa = 10.85DD274 pKa = 3.06KK275 pKa = 10.89RR276 pKa = 11.84LQEE279 pKa = 4.37SFF281 pKa = 3.6
MM1 pKa = 7.49CFEE4 pKa = 5.61PYY6 pKa = 9.17PLKK9 pKa = 11.04YY10 pKa = 10.22KK11 pKa = 10.22GAKK14 pKa = 9.28LHH16 pKa = 5.76LQGMTIPVPCGKK28 pKa = 10.13CPKK31 pKa = 8.62CTKK34 pKa = 10.46ARR36 pKa = 11.84INSWLFRR43 pKa = 11.84LTIHH47 pKa = 6.2ARR49 pKa = 11.84KK50 pKa = 9.52YY51 pKa = 9.66PYY53 pKa = 10.07PLFITLTYY61 pKa = 10.4DD62 pKa = 3.67DD63 pKa = 4.06KK64 pKa = 11.37HH65 pKa = 7.96LPVHH69 pKa = 6.3SKK71 pKa = 10.04SGRR74 pKa = 11.84VSLDD78 pKa = 2.85KK79 pKa = 10.99SDD81 pKa = 3.45PQKK84 pKa = 9.95WFKK87 pKa = 10.71RR88 pKa = 11.84LRR90 pKa = 11.84KK91 pKa = 9.97SLDD94 pKa = 3.15GQMDD98 pKa = 4.25LSYY101 pKa = 10.91FLVGEE106 pKa = 4.29YY107 pKa = 8.44GTKK110 pKa = 9.94RR111 pKa = 11.84KK112 pKa = 9.7RR113 pKa = 11.84PHH115 pKa = 5.26YY116 pKa = 10.13HH117 pKa = 4.97VLLYY121 pKa = 10.4GLPHH125 pKa = 6.45NKK127 pKa = 9.68SEE129 pKa = 5.46LIYY132 pKa = 9.24KK133 pKa = 8.33TWGLGFTMTLPVLDD147 pKa = 3.58GGMRR151 pKa = 11.84YY152 pKa = 8.56VLKK155 pKa = 11.14YY156 pKa = 8.09MAKK159 pKa = 9.53PRR161 pKa = 11.84SNYY164 pKa = 7.83TSKK167 pKa = 11.02LPEE170 pKa = 4.0FSLMSKK176 pKa = 10.71NIGANYY182 pKa = 7.71LTPQMIRR189 pKa = 11.84YY190 pKa = 8.67HH191 pKa = 8.25LEE193 pKa = 4.26DD194 pKa = 4.27PAHH197 pKa = 6.06TFASLGNGIKK207 pKa = 10.43LALPKK212 pKa = 10.33YY213 pKa = 8.45YY214 pKa = 9.97KK215 pKa = 10.46EE216 pKa = 4.44KK217 pKa = 10.63IFGIDD222 pKa = 2.86TPEE225 pKa = 4.01RR226 pKa = 11.84KK227 pKa = 9.3RR228 pKa = 11.84VTDD231 pKa = 3.58YY232 pKa = 10.61LQKK235 pKa = 10.09RR236 pKa = 11.84AEE238 pKa = 4.15MAQNSYY244 pKa = 10.4LRR246 pKa = 11.84RR247 pKa = 11.84LSLNNRR253 pKa = 11.84CLSQEE258 pKa = 4.25EE259 pKa = 4.37IIKK262 pKa = 10.49LAFLRR267 pKa = 11.84QHH269 pKa = 5.98SLSYY273 pKa = 10.85DD274 pKa = 3.06KK275 pKa = 10.89RR276 pKa = 11.84LQEE279 pKa = 4.37SFF281 pKa = 3.6
Molecular weight: 32.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1252 |
125 |
543 |
313.0 |
35.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.629 ± 0.818 | 0.639 ± 0.365 |
6.31 ± 1.12 | 4.872 ± 0.663 |
4.073 ± 0.452 | 6.629 ± 1.016 |
2.556 ± 0.875 | 5.112 ± 0.607 |
6.47 ± 1.44 | 9.824 ± 1.201 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.955 ± 0.511 | 5.751 ± 1.221 |
5.99 ± 0.928 | 4.633 ± 0.871 |
5.511 ± 0.594 | 6.15 ± 0.399 |
5.831 ± 0.742 | 4.313 ± 0.835 |
1.358 ± 0.403 | 4.393 ± 0.735 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
