
Amycolatopsis acidiphila
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Amycolatopsis
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
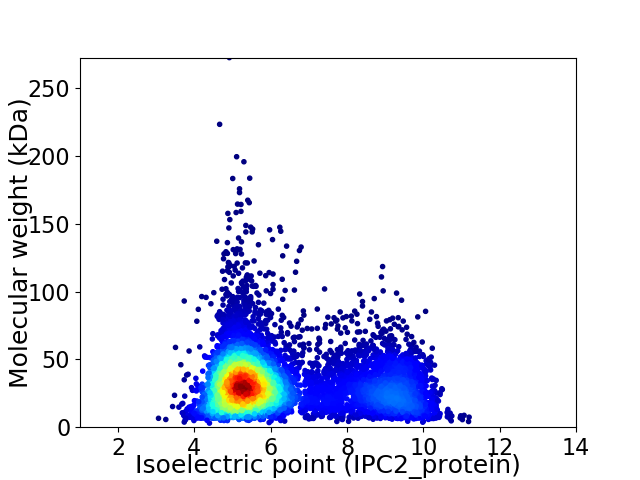
Virtual 2D-PAGE plot for 7527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A557ZZ67|A0A557ZZ67_9PSEU Uncharacterized protein OS=Amycolatopsis acidiphila OX=715473 GN=FNH06_31905 PE=4 SV=1
MM1 pKa = 7.16SQPIQTLHH9 pKa = 7.11DD10 pKa = 3.84FVLNLITDD18 pKa = 3.95DD19 pKa = 3.97SFGTSFLSDD28 pKa = 3.47PAGVLSGAGLGDD40 pKa = 3.51VTGLDD45 pKa = 3.7VQEE48 pKa = 4.39VTSLVADD55 pKa = 4.3HH56 pKa = 6.89LPAPVADD63 pKa = 3.53AVEE66 pKa = 4.52SGFAALPADD75 pKa = 3.65VTGVNDD81 pKa = 3.57LHH83 pKa = 6.89CALAHH88 pKa = 6.47LEE90 pKa = 4.16TVAAVAQDD98 pKa = 4.01LPVSVAGLSPSTTFAGDD115 pKa = 3.05AGGFTGAAALTGDD128 pKa = 4.69LVQTSGSLAGSTGGLAGGALADD150 pKa = 4.2TPAGAVTGAVAAATDD165 pKa = 4.06SVAAGVQSPLGTYY178 pKa = 10.04GVATDD183 pKa = 4.43SLPLSVPSFGSVSDD197 pKa = 3.97LGGTLDD203 pKa = 5.07SDD205 pKa = 4.28ALTHH209 pKa = 5.77GTSATAVAGGYY220 pKa = 8.35VASGGEE226 pKa = 3.8FAAAGIADD234 pKa = 3.96NSATLAGHH242 pKa = 7.17LSSVGDD248 pKa = 3.6VAAQPVAAGGDD259 pKa = 3.59EE260 pKa = 4.43LATHH264 pKa = 6.37VASGADD270 pKa = 3.34TLGDD274 pKa = 3.72VVSHH278 pKa = 6.06VPSVAAPAQLPADD291 pKa = 4.15LPVHH295 pKa = 6.57AVADD299 pKa = 4.65LPQSLPTPDD308 pKa = 3.66SVLPQVTHH316 pKa = 5.28TVEE319 pKa = 4.39STVSHH324 pKa = 6.82APLTDD329 pKa = 3.78SVSHH333 pKa = 6.53SSLIDD338 pKa = 3.65SVHH341 pKa = 6.24SALPDD346 pKa = 3.42VGDD349 pKa = 3.54LHH351 pKa = 7.01TDD353 pKa = 4.03LFHH356 pKa = 7.7GDD358 pKa = 3.95LPLGHH363 pKa = 7.28
MM1 pKa = 7.16SQPIQTLHH9 pKa = 7.11DD10 pKa = 3.84FVLNLITDD18 pKa = 3.95DD19 pKa = 3.97SFGTSFLSDD28 pKa = 3.47PAGVLSGAGLGDD40 pKa = 3.51VTGLDD45 pKa = 3.7VQEE48 pKa = 4.39VTSLVADD55 pKa = 4.3HH56 pKa = 6.89LPAPVADD63 pKa = 3.53AVEE66 pKa = 4.52SGFAALPADD75 pKa = 3.65VTGVNDD81 pKa = 3.57LHH83 pKa = 6.89CALAHH88 pKa = 6.47LEE90 pKa = 4.16TVAAVAQDD98 pKa = 4.01LPVSVAGLSPSTTFAGDD115 pKa = 3.05AGGFTGAAALTGDD128 pKa = 4.69LVQTSGSLAGSTGGLAGGALADD150 pKa = 4.2TPAGAVTGAVAAATDD165 pKa = 4.06SVAAGVQSPLGTYY178 pKa = 10.04GVATDD183 pKa = 4.43SLPLSVPSFGSVSDD197 pKa = 3.97LGGTLDD203 pKa = 5.07SDD205 pKa = 4.28ALTHH209 pKa = 5.77GTSATAVAGGYY220 pKa = 8.35VASGGEE226 pKa = 3.8FAAAGIADD234 pKa = 3.96NSATLAGHH242 pKa = 7.17LSSVGDD248 pKa = 3.6VAAQPVAAGGDD259 pKa = 3.59EE260 pKa = 4.43LATHH264 pKa = 6.37VASGADD270 pKa = 3.34TLGDD274 pKa = 3.72VVSHH278 pKa = 6.06VPSVAAPAQLPADD291 pKa = 4.15LPVHH295 pKa = 6.57AVADD299 pKa = 4.65LPQSLPTPDD308 pKa = 3.66SVLPQVTHH316 pKa = 5.28TVEE319 pKa = 4.39STVSHH324 pKa = 6.82APLTDD329 pKa = 3.78SVSHH333 pKa = 6.53SSLIDD338 pKa = 3.65SVHH341 pKa = 6.24SALPDD346 pKa = 3.42VGDD349 pKa = 3.54LHH351 pKa = 7.01TDD353 pKa = 4.03LFHH356 pKa = 7.7GDD358 pKa = 3.95LPLGHH363 pKa = 7.28
Molecular weight: 34.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A558AAR2|A0A558AAR2_9PSEU Alpha/beta hydrolase OS=Amycolatopsis acidiphila OX=715473 GN=FNH06_17405 PE=4 SV=1
MM1 pKa = 7.37GRR3 pKa = 11.84RR4 pKa = 11.84ARR6 pKa = 11.84AAAWRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ARR20 pKa = 11.84ARR22 pKa = 11.84PPRR25 pKa = 11.84PRR27 pKa = 11.84RR28 pKa = 11.84PTVSASPQGPPQPPRR43 pKa = 11.84SPRR46 pKa = 11.84SPQPPRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84DD55 pKa = 3.44PVTGRR60 pKa = 11.84PTMHH64 pKa = 7.18
MM1 pKa = 7.37GRR3 pKa = 11.84RR4 pKa = 11.84ARR6 pKa = 11.84AAAWRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ARR20 pKa = 11.84ARR22 pKa = 11.84PPRR25 pKa = 11.84PRR27 pKa = 11.84RR28 pKa = 11.84PTVSASPQGPPQPPRR43 pKa = 11.84SPRR46 pKa = 11.84SPQPPRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84DD55 pKa = 3.44PVTGRR60 pKa = 11.84PTMHH64 pKa = 7.18
Molecular weight: 7.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2361779 |
26 |
2563 |
313.8 |
33.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.076 ± 0.036 | 0.801 ± 0.009 |
5.815 ± 0.024 | 5.788 ± 0.025 |
2.969 ± 0.014 | 9.304 ± 0.024 |
2.231 ± 0.013 | 3.536 ± 0.017 |
2.126 ± 0.018 | 10.584 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.728 ± 0.01 | 1.928 ± 0.014 |
5.825 ± 0.02 | 2.948 ± 0.017 |
7.887 ± 0.032 | 5.125 ± 0.02 |
5.86 ± 0.023 | 8.93 ± 0.026 |
1.474 ± 0.011 | 2.065 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
