
Subsaximicrobium wynnwilliamsii
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Subsaximicrobium
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
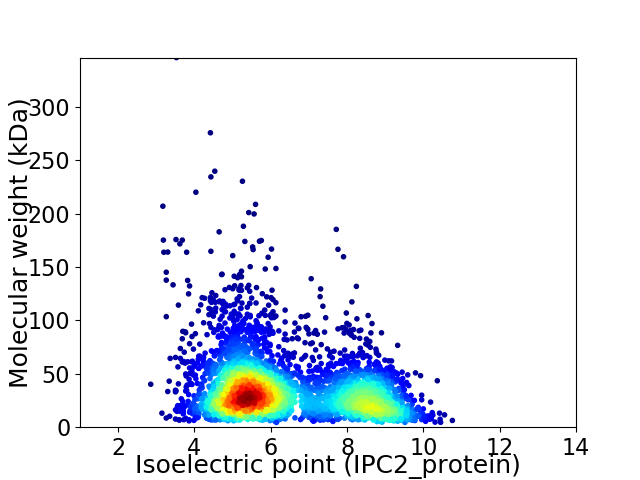
Virtual 2D-PAGE plot for 3955 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
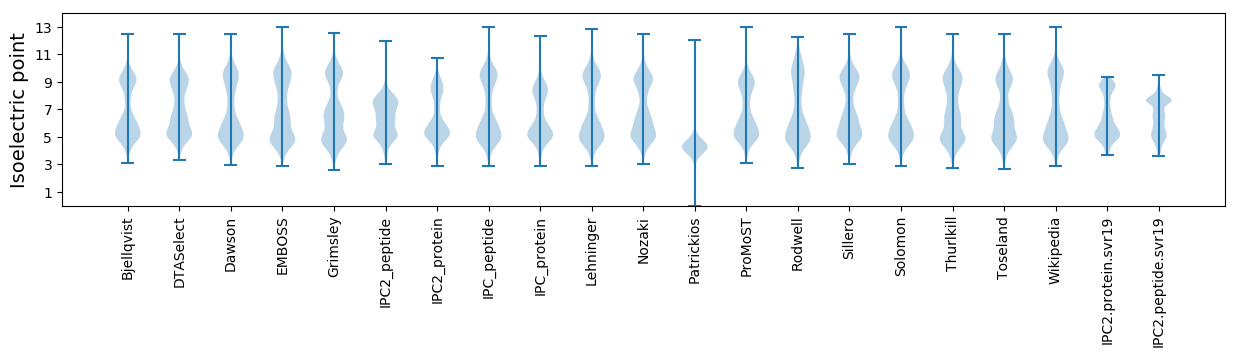
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C6ZEX4|A0A5C6ZEX4_9FLAO tRNA (Adenosine(37)-N6)-threonylcarbamoyltransferase complex dimerization subunit type 1 TsaB OS=Subsaximicrobium wynnwilliamsii OX=291179 GN=tsaB PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.36HH3 pKa = 5.51LTTLKK8 pKa = 10.87LIGFLILLLITSPVIHH24 pKa = 6.42AHH26 pKa = 6.19SLLSDD31 pKa = 4.22DD32 pKa = 5.18PLLDD36 pKa = 3.68TKK38 pKa = 11.55DD39 pKa = 5.19DD40 pKa = 3.69ITSSIDD46 pKa = 3.07PSVNVFIDD54 pKa = 3.42KK55 pKa = 10.78DD56 pKa = 3.94VLQAEE61 pKa = 4.43ALLNTTLNHH70 pKa = 7.14DD71 pKa = 4.38DD72 pKa = 4.02GVFHH76 pKa = 7.3LFTHH80 pKa = 6.65GKK82 pKa = 8.99PGQLLIDD89 pKa = 4.2GQWLQKK95 pKa = 10.69EE96 pKa = 4.48EE97 pKa = 3.96IVSFINSKK105 pKa = 10.74FNFPLGEE112 pKa = 3.98NSEE115 pKa = 4.3GQRR118 pKa = 11.84GLNIYY123 pKa = 9.6GCNFAQGEE131 pKa = 4.31KK132 pKa = 9.65GQKK135 pKa = 9.2AVAYY139 pKa = 10.05LEE141 pKa = 4.24KK142 pKa = 10.73QLGITIAASTNVTGKK157 pKa = 10.5DD158 pKa = 3.31GDD160 pKa = 3.76WQLEE164 pKa = 4.51SGTPTQLKK172 pKa = 10.17ALQNYY177 pKa = 9.12AYY179 pKa = 9.74NLQISCPNGLDD190 pKa = 3.16VHH192 pKa = 6.82IANDD196 pKa = 3.32VSGSVNSTEE205 pKa = 3.93FSQAKK210 pKa = 10.28DD211 pKa = 4.53FIAQLGASFDD221 pKa = 3.93NDD223 pKa = 3.1FGTLNTQSRR232 pKa = 11.84ISISNWSAGSNRR244 pKa = 11.84FVEE247 pKa = 4.27YY248 pKa = 10.33DD249 pKa = 3.78FPSAGQNYY257 pKa = 6.28TTSIADD263 pKa = 3.32ILSYY267 pKa = 10.73SSSSRR272 pKa = 11.84PFGGQTDD279 pKa = 4.35VYY281 pKa = 10.11TALLRR286 pKa = 11.84ANEE289 pKa = 4.23WVRR292 pKa = 11.84QNPVSDD298 pKa = 3.36RR299 pKa = 11.84DD300 pKa = 3.76VPKK303 pKa = 10.91VIVLLTDD310 pKa = 4.06ASCSQVANNISSLATQIKK328 pKa = 9.05NQGIYY333 pKa = 10.13IVVLAIDD340 pKa = 4.52DD341 pKa = 4.49AASCTALQGINVASAGGYY359 pKa = 9.79FSANNYY365 pKa = 7.57NTLQQDD371 pKa = 4.72AITFIQNIATASCDD385 pKa = 3.33NGGPAFFDD393 pKa = 3.61LTVSLSDD400 pKa = 5.65FVITDD405 pKa = 3.78CNTTAVASANYY416 pKa = 7.95TVSNISSGDD425 pKa = 3.52DD426 pKa = 3.42FNANLQVSFYY436 pKa = 10.87NGDD439 pKa = 3.32PTQPEE444 pKa = 3.99TQYY447 pKa = 11.36IFTDD451 pKa = 3.34NAGTQNLANGMGTYY465 pKa = 10.5SSTVNSDD472 pKa = 3.14ALINTSTLYY481 pKa = 11.02AVVNFDD487 pKa = 4.45GSLPGNAVPLSLSDD501 pKa = 4.48LSNQINVANEE511 pKa = 2.96GRR513 pKa = 11.84TFNNISTPVTRR524 pKa = 11.84DD525 pKa = 3.33DD526 pKa = 4.69GAGCQPFSTIDD537 pKa = 3.52VQVSNSGVGCDD548 pKa = 4.51DD549 pKa = 3.41EE550 pKa = 5.9VIYY553 pKa = 9.62TLQICNTGTADD564 pKa = 4.16ALMSPSDD571 pKa = 4.27IIPYY575 pKa = 10.05APAGFSLSSSTLIGTDD591 pKa = 3.18PFDD594 pKa = 3.91NLTGITASQLGQDD607 pKa = 3.72IDD609 pKa = 4.41GEE611 pKa = 4.32ALGDD615 pKa = 3.64GSGASVSMSADD626 pKa = 3.04GTTVAIGASANDD638 pKa = 3.59VNGSNSGHH646 pKa = 4.65VRR648 pKa = 11.84AYY650 pKa = 10.1HH651 pKa = 6.54LSGGTWNQRR660 pKa = 11.84GQDD663 pKa = 3.4IDD665 pKa = 3.95GEE667 pKa = 4.43AAFDD671 pKa = 3.53SSGFSVSMSADD682 pKa = 3.21GNTVAIGAIGTDD694 pKa = 3.81GNGSNSGHH702 pKa = 5.34VRR704 pKa = 11.84LYY706 pKa = 10.69DD707 pKa = 3.31WSGTAWIQRR716 pKa = 11.84GLDD719 pKa = 3.22IDD721 pKa = 4.35GEE723 pKa = 4.41AAGDD727 pKa = 3.6SSGFSVSMTSDD738 pKa = 3.1GNTLAIGARR747 pKa = 11.84NNDD750 pKa = 3.52GNTGIDD756 pKa = 3.38TDD758 pKa = 3.6NRR760 pKa = 11.84GHH762 pKa = 5.6VRR764 pKa = 11.84IYY766 pKa = 11.02DD767 pKa = 3.51WAGGSWNQRR776 pKa = 11.84GLDD779 pKa = 3.47IDD781 pKa = 4.29GEE783 pKa = 4.4AAGDD787 pKa = 3.58EE788 pKa = 4.74SGWSVSMSVDD798 pKa = 3.1GNTVAIGATDD808 pKa = 3.51NDD810 pKa = 4.66SNGSKK815 pKa = 9.95SGHH818 pKa = 4.23VRR820 pKa = 11.84IYY822 pKa = 10.7DD823 pKa = 3.4WSGTAWTQRR832 pKa = 11.84GLDD835 pKa = 3.25IDD837 pKa = 4.44GEE839 pKa = 4.27NSNDD843 pKa = 3.22EE844 pKa = 4.61SGWSVSISADD854 pKa = 3.25GNTVAIGAIDD864 pKa = 3.67NDD866 pKa = 4.19GGGGSSGHH874 pKa = 4.48VRR876 pKa = 11.84VYY878 pKa = 10.0EE879 pKa = 3.94WSGTAWTQRR888 pKa = 11.84GLDD891 pKa = 3.6IDD893 pKa = 5.48GEE895 pKa = 4.54DD896 pKa = 3.83NDD898 pKa = 5.34DD899 pKa = 3.52EE900 pKa = 5.01SGYY903 pKa = 9.83SVSMSADD910 pKa = 3.19GNTVATGAHH919 pKa = 6.2RR920 pKa = 11.84NDD922 pKa = 3.24GNAFRR927 pKa = 11.84AGHH930 pKa = 5.17VRR932 pKa = 11.84VYY934 pKa = 10.47DD935 pKa = 3.49WSGGSWTQRR944 pKa = 11.84GLDD947 pKa = 3.24IDD949 pKa = 4.4GEE951 pKa = 4.43AFGDD955 pKa = 3.75YY956 pKa = 10.26SGRR959 pKa = 11.84SVSMSADD966 pKa = 3.26GNTVAIGAPSNDD978 pKa = 3.25GNGTSSGHH986 pKa = 4.5VRR988 pKa = 11.84VYY990 pKa = 10.9SYY992 pKa = 11.86GSGAEE997 pKa = 3.95ILPASTCATYY1007 pKa = 10.34EE1008 pKa = 3.61YY1009 pKa = 9.46TYY1011 pKa = 10.8NITGVAATAGTPYY1024 pKa = 10.57DD1025 pKa = 3.53YY1026 pKa = 11.43SLAVNASAQSGNDD1039 pKa = 3.03PVTFTPNTNFSADD1052 pKa = 3.38GNTGLNGFDD1061 pKa = 3.22GAVNNTDD1068 pKa = 3.44DD1069 pKa = 3.77LTYY1072 pKa = 11.12DD1073 pKa = 3.94GVTTPCTAGDD1083 pKa = 3.79QIAVSVSMTGDD1094 pKa = 4.22GSCPGSFSTATVTVEE1109 pKa = 4.03NTSGTTISHH1118 pKa = 7.21ALLTLDD1124 pKa = 5.33LSGTDD1129 pKa = 2.82AFYY1132 pKa = 11.17NGEE1135 pKa = 4.47PYY1137 pKa = 11.01NFTNGMVLEE1146 pKa = 4.7EE1147 pKa = 4.72PNIFDD1152 pKa = 3.87PAYY1155 pKa = 9.08PAVPNALSNQTGSQQLAIFALPDD1178 pKa = 3.44GTSTFEE1184 pKa = 4.13LDD1186 pKa = 3.62VAVGTSVIDD1195 pKa = 3.96LSATIAQIPTNLNAGGSAIGSAGIAPAVEE1224 pKa = 4.44PSISGTPPGDD1234 pKa = 3.22VTTAATTIALSGYY1247 pKa = 9.75AATNATSVSWTSGSNGSFANASTASTTYY1275 pKa = 10.09TINDD1279 pKa = 3.05QDD1281 pKa = 3.26RR1282 pKa = 11.84AYY1284 pKa = 10.76GFVEE1288 pKa = 4.66LSLEE1292 pKa = 4.16ALSSGGCSTLTSFRR1306 pKa = 11.84VNITGGTYY1314 pKa = 10.1DD1315 pKa = 3.66YY1316 pKa = 11.67GDD1318 pKa = 3.94AQSSFDD1324 pKa = 3.71SGEE1327 pKa = 4.02TQLPVAGAALISNDD1341 pKa = 3.44VFLGTIPADD1350 pKa = 3.58AEE1352 pKa = 3.93AAAQPTADD1360 pKa = 3.76ATGDD1364 pKa = 3.72GADD1367 pKa = 3.75EE1368 pKa = 5.87DD1369 pKa = 4.59GLQTLTFGNFQPGNLYY1385 pKa = 9.04TLSVDD1390 pKa = 3.27ATNNSAALAYY1400 pKa = 9.81LHH1402 pKa = 7.27AYY1404 pKa = 8.96IDD1406 pKa = 3.81WNDD1409 pKa = 3.81DD1410 pKa = 2.94GDD1412 pKa = 4.43FIGDD1416 pKa = 3.63EE1417 pKa = 4.57GEE1419 pKa = 4.38KK1420 pKa = 10.57SLLVNVPAGSGAATYY1435 pKa = 8.78EE1436 pKa = 4.18VNFIIPSNGAAFTSSTIVRR1455 pKa = 11.84LRR1457 pKa = 11.84LSSDD1461 pKa = 2.83EE1462 pKa = 4.15FAAGRR1467 pKa = 11.84SYY1469 pKa = 11.01GAAANGEE1476 pKa = 4.2IEE1478 pKa = 5.46DD1479 pKa = 4.06FLIDD1483 pKa = 3.41EE1484 pKa = 4.53TLSVEE1489 pKa = 3.95NLEE1492 pKa = 4.45AKK1494 pKa = 10.37GISIYY1499 pKa = 10.32PNPASDD1505 pKa = 4.39FLIIDD1510 pKa = 4.21FGQNAGRR1517 pKa = 11.84FNTVEE1522 pKa = 4.48LYY1524 pKa = 10.64DD1525 pKa = 3.24ITGRR1529 pKa = 11.84LMSKK1533 pKa = 9.42TSLKK1537 pKa = 10.47EE1538 pKa = 3.77NPLDD1542 pKa = 3.69KK1543 pKa = 11.0VQVRR1547 pKa = 11.84IDD1549 pKa = 3.39QLNAGIYY1556 pKa = 10.18LLNLSSKK1563 pKa = 10.3EE1564 pKa = 3.93DD1565 pKa = 3.4QFSQRR1570 pKa = 11.84VIIRR1574 pKa = 3.56
MM1 pKa = 7.42KK2 pKa = 10.36HH3 pKa = 5.51LTTLKK8 pKa = 10.87LIGFLILLLITSPVIHH24 pKa = 6.42AHH26 pKa = 6.19SLLSDD31 pKa = 4.22DD32 pKa = 5.18PLLDD36 pKa = 3.68TKK38 pKa = 11.55DD39 pKa = 5.19DD40 pKa = 3.69ITSSIDD46 pKa = 3.07PSVNVFIDD54 pKa = 3.42KK55 pKa = 10.78DD56 pKa = 3.94VLQAEE61 pKa = 4.43ALLNTTLNHH70 pKa = 7.14DD71 pKa = 4.38DD72 pKa = 4.02GVFHH76 pKa = 7.3LFTHH80 pKa = 6.65GKK82 pKa = 8.99PGQLLIDD89 pKa = 4.2GQWLQKK95 pKa = 10.69EE96 pKa = 4.48EE97 pKa = 3.96IVSFINSKK105 pKa = 10.74FNFPLGEE112 pKa = 3.98NSEE115 pKa = 4.3GQRR118 pKa = 11.84GLNIYY123 pKa = 9.6GCNFAQGEE131 pKa = 4.31KK132 pKa = 9.65GQKK135 pKa = 9.2AVAYY139 pKa = 10.05LEE141 pKa = 4.24KK142 pKa = 10.73QLGITIAASTNVTGKK157 pKa = 10.5DD158 pKa = 3.31GDD160 pKa = 3.76WQLEE164 pKa = 4.51SGTPTQLKK172 pKa = 10.17ALQNYY177 pKa = 9.12AYY179 pKa = 9.74NLQISCPNGLDD190 pKa = 3.16VHH192 pKa = 6.82IANDD196 pKa = 3.32VSGSVNSTEE205 pKa = 3.93FSQAKK210 pKa = 10.28DD211 pKa = 4.53FIAQLGASFDD221 pKa = 3.93NDD223 pKa = 3.1FGTLNTQSRR232 pKa = 11.84ISISNWSAGSNRR244 pKa = 11.84FVEE247 pKa = 4.27YY248 pKa = 10.33DD249 pKa = 3.78FPSAGQNYY257 pKa = 6.28TTSIADD263 pKa = 3.32ILSYY267 pKa = 10.73SSSSRR272 pKa = 11.84PFGGQTDD279 pKa = 4.35VYY281 pKa = 10.11TALLRR286 pKa = 11.84ANEE289 pKa = 4.23WVRR292 pKa = 11.84QNPVSDD298 pKa = 3.36RR299 pKa = 11.84DD300 pKa = 3.76VPKK303 pKa = 10.91VIVLLTDD310 pKa = 4.06ASCSQVANNISSLATQIKK328 pKa = 9.05NQGIYY333 pKa = 10.13IVVLAIDD340 pKa = 4.52DD341 pKa = 4.49AASCTALQGINVASAGGYY359 pKa = 9.79FSANNYY365 pKa = 7.57NTLQQDD371 pKa = 4.72AITFIQNIATASCDD385 pKa = 3.33NGGPAFFDD393 pKa = 3.61LTVSLSDD400 pKa = 5.65FVITDD405 pKa = 3.78CNTTAVASANYY416 pKa = 7.95TVSNISSGDD425 pKa = 3.52DD426 pKa = 3.42FNANLQVSFYY436 pKa = 10.87NGDD439 pKa = 3.32PTQPEE444 pKa = 3.99TQYY447 pKa = 11.36IFTDD451 pKa = 3.34NAGTQNLANGMGTYY465 pKa = 10.5SSTVNSDD472 pKa = 3.14ALINTSTLYY481 pKa = 11.02AVVNFDD487 pKa = 4.45GSLPGNAVPLSLSDD501 pKa = 4.48LSNQINVANEE511 pKa = 2.96GRR513 pKa = 11.84TFNNISTPVTRR524 pKa = 11.84DD525 pKa = 3.33DD526 pKa = 4.69GAGCQPFSTIDD537 pKa = 3.52VQVSNSGVGCDD548 pKa = 4.51DD549 pKa = 3.41EE550 pKa = 5.9VIYY553 pKa = 9.62TLQICNTGTADD564 pKa = 4.16ALMSPSDD571 pKa = 4.27IIPYY575 pKa = 10.05APAGFSLSSSTLIGTDD591 pKa = 3.18PFDD594 pKa = 3.91NLTGITASQLGQDD607 pKa = 3.72IDD609 pKa = 4.41GEE611 pKa = 4.32ALGDD615 pKa = 3.64GSGASVSMSADD626 pKa = 3.04GTTVAIGASANDD638 pKa = 3.59VNGSNSGHH646 pKa = 4.65VRR648 pKa = 11.84AYY650 pKa = 10.1HH651 pKa = 6.54LSGGTWNQRR660 pKa = 11.84GQDD663 pKa = 3.4IDD665 pKa = 3.95GEE667 pKa = 4.43AAFDD671 pKa = 3.53SSGFSVSMSADD682 pKa = 3.21GNTVAIGAIGTDD694 pKa = 3.81GNGSNSGHH702 pKa = 5.34VRR704 pKa = 11.84LYY706 pKa = 10.69DD707 pKa = 3.31WSGTAWIQRR716 pKa = 11.84GLDD719 pKa = 3.22IDD721 pKa = 4.35GEE723 pKa = 4.41AAGDD727 pKa = 3.6SSGFSVSMTSDD738 pKa = 3.1GNTLAIGARR747 pKa = 11.84NNDD750 pKa = 3.52GNTGIDD756 pKa = 3.38TDD758 pKa = 3.6NRR760 pKa = 11.84GHH762 pKa = 5.6VRR764 pKa = 11.84IYY766 pKa = 11.02DD767 pKa = 3.51WAGGSWNQRR776 pKa = 11.84GLDD779 pKa = 3.47IDD781 pKa = 4.29GEE783 pKa = 4.4AAGDD787 pKa = 3.58EE788 pKa = 4.74SGWSVSMSVDD798 pKa = 3.1GNTVAIGATDD808 pKa = 3.51NDD810 pKa = 4.66SNGSKK815 pKa = 9.95SGHH818 pKa = 4.23VRR820 pKa = 11.84IYY822 pKa = 10.7DD823 pKa = 3.4WSGTAWTQRR832 pKa = 11.84GLDD835 pKa = 3.25IDD837 pKa = 4.44GEE839 pKa = 4.27NSNDD843 pKa = 3.22EE844 pKa = 4.61SGWSVSISADD854 pKa = 3.25GNTVAIGAIDD864 pKa = 3.67NDD866 pKa = 4.19GGGGSSGHH874 pKa = 4.48VRR876 pKa = 11.84VYY878 pKa = 10.0EE879 pKa = 3.94WSGTAWTQRR888 pKa = 11.84GLDD891 pKa = 3.6IDD893 pKa = 5.48GEE895 pKa = 4.54DD896 pKa = 3.83NDD898 pKa = 5.34DD899 pKa = 3.52EE900 pKa = 5.01SGYY903 pKa = 9.83SVSMSADD910 pKa = 3.19GNTVATGAHH919 pKa = 6.2RR920 pKa = 11.84NDD922 pKa = 3.24GNAFRR927 pKa = 11.84AGHH930 pKa = 5.17VRR932 pKa = 11.84VYY934 pKa = 10.47DD935 pKa = 3.49WSGGSWTQRR944 pKa = 11.84GLDD947 pKa = 3.24IDD949 pKa = 4.4GEE951 pKa = 4.43AFGDD955 pKa = 3.75YY956 pKa = 10.26SGRR959 pKa = 11.84SVSMSADD966 pKa = 3.26GNTVAIGAPSNDD978 pKa = 3.25GNGTSSGHH986 pKa = 4.5VRR988 pKa = 11.84VYY990 pKa = 10.9SYY992 pKa = 11.86GSGAEE997 pKa = 3.95ILPASTCATYY1007 pKa = 10.34EE1008 pKa = 3.61YY1009 pKa = 9.46TYY1011 pKa = 10.8NITGVAATAGTPYY1024 pKa = 10.57DD1025 pKa = 3.53YY1026 pKa = 11.43SLAVNASAQSGNDD1039 pKa = 3.03PVTFTPNTNFSADD1052 pKa = 3.38GNTGLNGFDD1061 pKa = 3.22GAVNNTDD1068 pKa = 3.44DD1069 pKa = 3.77LTYY1072 pKa = 11.12DD1073 pKa = 3.94GVTTPCTAGDD1083 pKa = 3.79QIAVSVSMTGDD1094 pKa = 4.22GSCPGSFSTATVTVEE1109 pKa = 4.03NTSGTTISHH1118 pKa = 7.21ALLTLDD1124 pKa = 5.33LSGTDD1129 pKa = 2.82AFYY1132 pKa = 11.17NGEE1135 pKa = 4.47PYY1137 pKa = 11.01NFTNGMVLEE1146 pKa = 4.7EE1147 pKa = 4.72PNIFDD1152 pKa = 3.87PAYY1155 pKa = 9.08PAVPNALSNQTGSQQLAIFALPDD1178 pKa = 3.44GTSTFEE1184 pKa = 4.13LDD1186 pKa = 3.62VAVGTSVIDD1195 pKa = 3.96LSATIAQIPTNLNAGGSAIGSAGIAPAVEE1224 pKa = 4.44PSISGTPPGDD1234 pKa = 3.22VTTAATTIALSGYY1247 pKa = 9.75AATNATSVSWTSGSNGSFANASTASTTYY1275 pKa = 10.09TINDD1279 pKa = 3.05QDD1281 pKa = 3.26RR1282 pKa = 11.84AYY1284 pKa = 10.76GFVEE1288 pKa = 4.66LSLEE1292 pKa = 4.16ALSSGGCSTLTSFRR1306 pKa = 11.84VNITGGTYY1314 pKa = 10.1DD1315 pKa = 3.66YY1316 pKa = 11.67GDD1318 pKa = 3.94AQSSFDD1324 pKa = 3.71SGEE1327 pKa = 4.02TQLPVAGAALISNDD1341 pKa = 3.44VFLGTIPADD1350 pKa = 3.58AEE1352 pKa = 3.93AAAQPTADD1360 pKa = 3.76ATGDD1364 pKa = 3.72GADD1367 pKa = 3.75EE1368 pKa = 5.87DD1369 pKa = 4.59GLQTLTFGNFQPGNLYY1385 pKa = 9.04TLSVDD1390 pKa = 3.27ATNNSAALAYY1400 pKa = 9.81LHH1402 pKa = 7.27AYY1404 pKa = 8.96IDD1406 pKa = 3.81WNDD1409 pKa = 3.81DD1410 pKa = 2.94GDD1412 pKa = 4.43FIGDD1416 pKa = 3.63EE1417 pKa = 4.57GEE1419 pKa = 4.38KK1420 pKa = 10.57SLLVNVPAGSGAATYY1435 pKa = 8.78EE1436 pKa = 4.18VNFIIPSNGAAFTSSTIVRR1455 pKa = 11.84LRR1457 pKa = 11.84LSSDD1461 pKa = 2.83EE1462 pKa = 4.15FAAGRR1467 pKa = 11.84SYY1469 pKa = 11.01GAAANGEE1476 pKa = 4.2IEE1478 pKa = 5.46DD1479 pKa = 4.06FLIDD1483 pKa = 3.41EE1484 pKa = 4.53TLSVEE1489 pKa = 3.95NLEE1492 pKa = 4.45AKK1494 pKa = 10.37GISIYY1499 pKa = 10.32PNPASDD1505 pKa = 4.39FLIIDD1510 pKa = 4.21FGQNAGRR1517 pKa = 11.84FNTVEE1522 pKa = 4.48LYY1524 pKa = 10.64DD1525 pKa = 3.24ITGRR1529 pKa = 11.84LMSKK1533 pKa = 9.42TSLKK1537 pKa = 10.47EE1538 pKa = 3.77NPLDD1542 pKa = 3.69KK1543 pKa = 11.0VQVRR1547 pKa = 11.84IDD1549 pKa = 3.39QLNAGIYY1556 pKa = 10.18LLNLSSKK1563 pKa = 10.3EE1564 pKa = 3.93DD1565 pKa = 3.4QFSQRR1570 pKa = 11.84VIIRR1574 pKa = 3.56
Molecular weight: 163.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C6ZE19|A0A5C6ZE19_9FLAO Xanthine dehydrogenase family protein molybdopterin-binding subunit OS=Subsaximicrobium wynnwilliamsii OX=291179 GN=ESY86_13270 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.05GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.41VSVSSEE48 pKa = 3.83VRR50 pKa = 11.84HH51 pKa = 6.04KK52 pKa = 10.77KK53 pKa = 10.01
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.05GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.41VSVSSEE48 pKa = 3.83VRR50 pKa = 11.84HH51 pKa = 6.04KK52 pKa = 10.77KK53 pKa = 10.01
Molecular weight: 6.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1316798 |
37 |
3305 |
332.9 |
37.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.762 ± 0.045 | 0.757 ± 0.014 |
5.814 ± 0.034 | 6.589 ± 0.041 |
5.293 ± 0.039 | 6.203 ± 0.045 |
1.803 ± 0.021 | 7.718 ± 0.038 |
7.463 ± 0.054 | 9.434 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.021 | 6.062 ± 0.041 |
3.299 ± 0.022 | 3.595 ± 0.023 |
3.467 ± 0.029 | 6.67 ± 0.031 |
5.784 ± 0.045 | 6.039 ± 0.031 |
1.011 ± 0.015 | 3.981 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
