
Methanofollis sp. FWC-SCC2
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanomicrobiales; Methanomicrobiaceae; Methanofollis; unclassified Methanofollis
Average proteome isoelectric point is 5.75
Get precalculated fractions of proteins
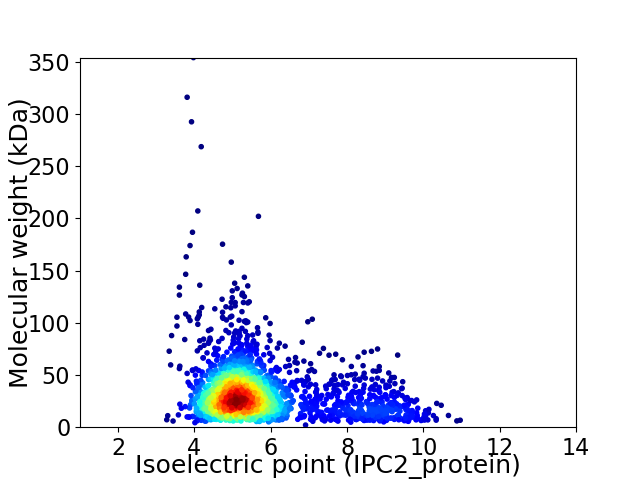
Virtual 2D-PAGE plot for 2257 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A483CQC9|A0A483CQC9_9EURY Uncharacterized protein OS=Methanofollis sp. FWC-SCC2 OX=2052832 GN=CUJ86_06325 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 9.44MHH4 pKa = 6.59SVLAAFVLVLFFCQAATASDD24 pKa = 4.01SPEE27 pKa = 3.89VTISADD33 pKa = 3.77PLWSVAGDD41 pKa = 3.53GGSVSLHH48 pKa = 5.42VLVADD53 pKa = 4.83GGAGVNAAAVDD64 pKa = 4.4LSCDD68 pKa = 3.9PSCGTITPAHH78 pKa = 6.78PVTNAHH84 pKa = 5.65GTATATFHH92 pKa = 7.07PSTVAGPVEE101 pKa = 4.27ITASAIWDD109 pKa = 3.97GQDD112 pKa = 3.3VPVTGSVVVHH122 pKa = 6.97IDD124 pKa = 3.06HH125 pKa = 7.95DD126 pKa = 4.16DD127 pKa = 3.98PYY129 pKa = 11.48AITLLEE135 pKa = 4.38FPEE138 pKa = 4.89GATVGSDD145 pKa = 3.08VTITVGIEE153 pKa = 4.0DD154 pKa = 3.94CWGNPIDD161 pKa = 5.28DD162 pKa = 4.63ALDD165 pKa = 3.66VEE167 pKa = 4.96TVNLSVGSPGNAAAFVTDD185 pKa = 4.55GGTGDD190 pKa = 5.87AITLPLDD197 pKa = 3.66SAGQAAATLRR207 pKa = 11.84LDD209 pKa = 3.42TTAGEE214 pKa = 4.42NIVFLDD220 pKa = 3.88APGTVADD227 pKa = 3.92VYY229 pKa = 8.99MTIEE233 pKa = 4.36GVANGTPVSITSAYY247 pKa = 10.5ASASCPANGEE257 pKa = 4.13DD258 pKa = 3.96TVLIAYY264 pKa = 9.56SLLDD268 pKa = 3.58TYY270 pKa = 11.73GNPCSGADD278 pKa = 3.46VRR280 pKa = 11.84IASSLGEE287 pKa = 4.2TATVATNRR295 pKa = 11.84FGSALVRR302 pKa = 11.84YY303 pKa = 8.35GPKK306 pKa = 8.8VTPVDD311 pKa = 3.79VVLTATAVGNGSVTTSDD328 pKa = 3.29TVSFTSTAPVNMALTASLLNMKK350 pKa = 10.18SLDD353 pKa = 3.57VDD355 pKa = 4.01SSSTAAVTARR365 pKa = 11.84ITTEE369 pKa = 3.79GGTPPDD375 pKa = 3.73EE376 pKa = 4.15EE377 pKa = 4.82VEE379 pKa = 4.58VTFSITDD386 pKa = 3.41EE387 pKa = 4.05EE388 pKa = 4.87RR389 pKa = 11.84GGGTAPSLNLATVTTSGGVATATFIPGEE417 pKa = 4.21FSSSDD422 pKa = 3.64PSTGTCTLTATATLNGTPVTRR443 pKa = 11.84SLDD446 pKa = 3.89LAWRR450 pKa = 11.84NYY452 pKa = 9.1PSVSVSVSAEE462 pKa = 3.83PEE464 pKa = 4.05TLVVNNTADD473 pKa = 3.37VTISIRR479 pKa = 11.84GDD481 pKa = 2.85GWEE484 pKa = 4.37FAPPPADD491 pKa = 2.96VMMCISRR498 pKa = 11.84TPTMLRR504 pKa = 11.84GYY506 pKa = 8.77PDD508 pKa = 3.47PMVIAMNASMYY519 pKa = 10.42FAGLMDD525 pKa = 4.81EE526 pKa = 5.64DD527 pKa = 4.32VDD529 pKa = 4.44SLGLVSYY536 pKa = 11.19GEE538 pKa = 4.09EE539 pKa = 4.17TPAGLMADD547 pKa = 3.78LRR549 pKa = 11.84EE550 pKa = 4.23MNSGNTKK557 pKa = 10.07SVYY560 pKa = 10.59NEE562 pKa = 3.39IGYY565 pKa = 10.22DD566 pKa = 3.43PGGGSGAEE574 pKa = 4.09AYY576 pKa = 7.73ITTHH580 pKa = 4.68YY581 pKa = 8.2TKK583 pKa = 10.58SGTVRR588 pKa = 11.84FDD590 pKa = 3.62EE591 pKa = 4.82EE592 pKa = 4.4VTLDD596 pKa = 3.87LPLSTSYY603 pKa = 11.66GSFNDD608 pKa = 3.84SVQCLIPYY616 pKa = 10.11SSLSKK621 pKa = 10.92GQGKK625 pKa = 9.78HH626 pKa = 6.39SGDD629 pKa = 3.82EE630 pKa = 4.03LRR632 pKa = 11.84LALYY636 pKa = 10.38KK637 pKa = 10.73SITEE641 pKa = 3.97IVDD644 pKa = 3.02NGSADD649 pKa = 3.56DD650 pKa = 3.78ARR652 pKa = 11.84AVVVLLDD659 pKa = 4.42DD660 pKa = 4.55FWNNGGGGKK669 pKa = 10.36FGTLYY674 pKa = 10.92GNPLGPDD681 pKa = 3.18GTYY684 pKa = 10.56EE685 pKa = 4.23KK686 pKa = 10.15EE687 pKa = 4.01DD688 pKa = 3.99YY689 pKa = 11.14YY690 pKa = 10.58IFPDD694 pKa = 3.66DD695 pKa = 3.69GRR697 pKa = 11.84GNGHH701 pKa = 5.48MWGSGGRR708 pKa = 11.84WQNLANYY715 pKa = 10.27ASDD718 pKa = 3.56NDD720 pKa = 3.38VRR722 pKa = 11.84IYY724 pKa = 11.22VLLIDD729 pKa = 3.73THH731 pKa = 6.77NGNVNGKK738 pKa = 9.31IGRR741 pKa = 11.84DD742 pKa = 3.38LSTLADD748 pKa = 3.64QTDD751 pKa = 3.48GEE753 pKa = 4.54YY754 pKa = 10.65CYY756 pKa = 11.15VEE758 pKa = 4.94NEE760 pKa = 4.29DD761 pKa = 4.79FEE763 pKa = 6.72SDD765 pKa = 3.86LEE767 pKa = 4.63GFLEE771 pKa = 5.62DD772 pKa = 4.04IKK774 pKa = 11.55DD775 pKa = 3.93DD776 pKa = 3.86LRR778 pKa = 11.84VASVFNTALDD788 pKa = 3.55ASLADD793 pKa = 3.38VTVDD797 pKa = 3.58GEE799 pKa = 4.87SVSSAGVLSYY809 pKa = 10.77EE810 pKa = 4.19YY811 pKa = 11.2VEE813 pKa = 6.11GVSTWITNTSATGDD827 pKa = 3.8DD828 pKa = 4.72LYY830 pKa = 11.48SGTVDD835 pKa = 5.18DD836 pKa = 4.61TGDD839 pKa = 3.37WADD842 pKa = 4.16DD843 pKa = 3.5RR844 pKa = 11.84VLSFGIGTIEE854 pKa = 4.05YY855 pKa = 8.34NQVWTATVRR864 pKa = 11.84LNFSSAGSIGLFTDD878 pKa = 3.56EE879 pKa = 4.88NSVISLSDD887 pKa = 3.52GEE889 pKa = 4.53GNDD892 pKa = 3.81YY893 pKa = 11.1EE894 pKa = 4.76QVLPPLSISVVDD906 pKa = 4.34PDD908 pKa = 5.23DD909 pKa = 3.98VGAFNTTAFDD919 pKa = 3.63VTYY922 pKa = 8.66LTPTGNNEE930 pKa = 3.85TGSIEE935 pKa = 4.3VEE937 pKa = 3.31WGLAYY942 pKa = 10.21EE943 pKa = 4.61GANPEE948 pKa = 4.55SVTQHH953 pKa = 6.52LSYY956 pKa = 10.59QYY958 pKa = 11.27SPDD961 pKa = 3.63NIVWDD966 pKa = 4.15NVWIPVPDD974 pKa = 3.54IPAVYY979 pKa = 9.74EE980 pKa = 4.23GVDD983 pKa = 3.65GVYY986 pKa = 10.59SADD989 pKa = 3.9FSTSGNNGWYY999 pKa = 9.95KK1000 pKa = 9.71IRR1002 pKa = 11.84VRR1004 pKa = 11.84ARR1006 pKa = 11.84EE1007 pKa = 4.28AIDD1010 pKa = 3.64GGAEE1014 pKa = 3.6AWRR1017 pKa = 11.84TADD1020 pKa = 3.62YY1021 pKa = 11.0LVLVNSGNKK1030 pKa = 9.73LVMRR1034 pKa = 11.84ILL1036 pKa = 3.73
MM1 pKa = 7.5KK2 pKa = 9.44MHH4 pKa = 6.59SVLAAFVLVLFFCQAATASDD24 pKa = 4.01SPEE27 pKa = 3.89VTISADD33 pKa = 3.77PLWSVAGDD41 pKa = 3.53GGSVSLHH48 pKa = 5.42VLVADD53 pKa = 4.83GGAGVNAAAVDD64 pKa = 4.4LSCDD68 pKa = 3.9PSCGTITPAHH78 pKa = 6.78PVTNAHH84 pKa = 5.65GTATATFHH92 pKa = 7.07PSTVAGPVEE101 pKa = 4.27ITASAIWDD109 pKa = 3.97GQDD112 pKa = 3.3VPVTGSVVVHH122 pKa = 6.97IDD124 pKa = 3.06HH125 pKa = 7.95DD126 pKa = 4.16DD127 pKa = 3.98PYY129 pKa = 11.48AITLLEE135 pKa = 4.38FPEE138 pKa = 4.89GATVGSDD145 pKa = 3.08VTITVGIEE153 pKa = 4.0DD154 pKa = 3.94CWGNPIDD161 pKa = 5.28DD162 pKa = 4.63ALDD165 pKa = 3.66VEE167 pKa = 4.96TVNLSVGSPGNAAAFVTDD185 pKa = 4.55GGTGDD190 pKa = 5.87AITLPLDD197 pKa = 3.66SAGQAAATLRR207 pKa = 11.84LDD209 pKa = 3.42TTAGEE214 pKa = 4.42NIVFLDD220 pKa = 3.88APGTVADD227 pKa = 3.92VYY229 pKa = 8.99MTIEE233 pKa = 4.36GVANGTPVSITSAYY247 pKa = 10.5ASASCPANGEE257 pKa = 4.13DD258 pKa = 3.96TVLIAYY264 pKa = 9.56SLLDD268 pKa = 3.58TYY270 pKa = 11.73GNPCSGADD278 pKa = 3.46VRR280 pKa = 11.84IASSLGEE287 pKa = 4.2TATVATNRR295 pKa = 11.84FGSALVRR302 pKa = 11.84YY303 pKa = 8.35GPKK306 pKa = 8.8VTPVDD311 pKa = 3.79VVLTATAVGNGSVTTSDD328 pKa = 3.29TVSFTSTAPVNMALTASLLNMKK350 pKa = 10.18SLDD353 pKa = 3.57VDD355 pKa = 4.01SSSTAAVTARR365 pKa = 11.84ITTEE369 pKa = 3.79GGTPPDD375 pKa = 3.73EE376 pKa = 4.15EE377 pKa = 4.82VEE379 pKa = 4.58VTFSITDD386 pKa = 3.41EE387 pKa = 4.05EE388 pKa = 4.87RR389 pKa = 11.84GGGTAPSLNLATVTTSGGVATATFIPGEE417 pKa = 4.21FSSSDD422 pKa = 3.64PSTGTCTLTATATLNGTPVTRR443 pKa = 11.84SLDD446 pKa = 3.89LAWRR450 pKa = 11.84NYY452 pKa = 9.1PSVSVSVSAEE462 pKa = 3.83PEE464 pKa = 4.05TLVVNNTADD473 pKa = 3.37VTISIRR479 pKa = 11.84GDD481 pKa = 2.85GWEE484 pKa = 4.37FAPPPADD491 pKa = 2.96VMMCISRR498 pKa = 11.84TPTMLRR504 pKa = 11.84GYY506 pKa = 8.77PDD508 pKa = 3.47PMVIAMNASMYY519 pKa = 10.42FAGLMDD525 pKa = 4.81EE526 pKa = 5.64DD527 pKa = 4.32VDD529 pKa = 4.44SLGLVSYY536 pKa = 11.19GEE538 pKa = 4.09EE539 pKa = 4.17TPAGLMADD547 pKa = 3.78LRR549 pKa = 11.84EE550 pKa = 4.23MNSGNTKK557 pKa = 10.07SVYY560 pKa = 10.59NEE562 pKa = 3.39IGYY565 pKa = 10.22DD566 pKa = 3.43PGGGSGAEE574 pKa = 4.09AYY576 pKa = 7.73ITTHH580 pKa = 4.68YY581 pKa = 8.2TKK583 pKa = 10.58SGTVRR588 pKa = 11.84FDD590 pKa = 3.62EE591 pKa = 4.82EE592 pKa = 4.4VTLDD596 pKa = 3.87LPLSTSYY603 pKa = 11.66GSFNDD608 pKa = 3.84SVQCLIPYY616 pKa = 10.11SSLSKK621 pKa = 10.92GQGKK625 pKa = 9.78HH626 pKa = 6.39SGDD629 pKa = 3.82EE630 pKa = 4.03LRR632 pKa = 11.84LALYY636 pKa = 10.38KK637 pKa = 10.73SITEE641 pKa = 3.97IVDD644 pKa = 3.02NGSADD649 pKa = 3.56DD650 pKa = 3.78ARR652 pKa = 11.84AVVVLLDD659 pKa = 4.42DD660 pKa = 4.55FWNNGGGGKK669 pKa = 10.36FGTLYY674 pKa = 10.92GNPLGPDD681 pKa = 3.18GTYY684 pKa = 10.56EE685 pKa = 4.23KK686 pKa = 10.15EE687 pKa = 4.01DD688 pKa = 3.99YY689 pKa = 11.14YY690 pKa = 10.58IFPDD694 pKa = 3.66DD695 pKa = 3.69GRR697 pKa = 11.84GNGHH701 pKa = 5.48MWGSGGRR708 pKa = 11.84WQNLANYY715 pKa = 10.27ASDD718 pKa = 3.56NDD720 pKa = 3.38VRR722 pKa = 11.84IYY724 pKa = 11.22VLLIDD729 pKa = 3.73THH731 pKa = 6.77NGNVNGKK738 pKa = 9.31IGRR741 pKa = 11.84DD742 pKa = 3.38LSTLADD748 pKa = 3.64QTDD751 pKa = 3.48GEE753 pKa = 4.54YY754 pKa = 10.65CYY756 pKa = 11.15VEE758 pKa = 4.94NEE760 pKa = 4.29DD761 pKa = 4.79FEE763 pKa = 6.72SDD765 pKa = 3.86LEE767 pKa = 4.63GFLEE771 pKa = 5.62DD772 pKa = 4.04IKK774 pKa = 11.55DD775 pKa = 3.93DD776 pKa = 3.86LRR778 pKa = 11.84VASVFNTALDD788 pKa = 3.55ASLADD793 pKa = 3.38VTVDD797 pKa = 3.58GEE799 pKa = 4.87SVSSAGVLSYY809 pKa = 10.77EE810 pKa = 4.19YY811 pKa = 11.2VEE813 pKa = 6.11GVSTWITNTSATGDD827 pKa = 3.8DD828 pKa = 4.72LYY830 pKa = 11.48SGTVDD835 pKa = 5.18DD836 pKa = 4.61TGDD839 pKa = 3.37WADD842 pKa = 4.16DD843 pKa = 3.5RR844 pKa = 11.84VLSFGIGTIEE854 pKa = 4.05YY855 pKa = 8.34NQVWTATVRR864 pKa = 11.84LNFSSAGSIGLFTDD878 pKa = 3.56EE879 pKa = 4.88NSVISLSDD887 pKa = 3.52GEE889 pKa = 4.53GNDD892 pKa = 3.81YY893 pKa = 11.1EE894 pKa = 4.76QVLPPLSISVVDD906 pKa = 4.34PDD908 pKa = 5.23DD909 pKa = 3.98VGAFNTTAFDD919 pKa = 3.63VTYY922 pKa = 8.66LTPTGNNEE930 pKa = 3.85TGSIEE935 pKa = 4.3VEE937 pKa = 3.31WGLAYY942 pKa = 10.21EE943 pKa = 4.61GANPEE948 pKa = 4.55SVTQHH953 pKa = 6.52LSYY956 pKa = 10.59QYY958 pKa = 11.27SPDD961 pKa = 3.63NIVWDD966 pKa = 4.15NVWIPVPDD974 pKa = 3.54IPAVYY979 pKa = 9.74EE980 pKa = 4.23GVDD983 pKa = 3.65GVYY986 pKa = 10.59SADD989 pKa = 3.9FSTSGNNGWYY999 pKa = 9.95KK1000 pKa = 9.71IRR1002 pKa = 11.84VRR1004 pKa = 11.84ARR1006 pKa = 11.84EE1007 pKa = 4.28AIDD1010 pKa = 3.64GGAEE1014 pKa = 3.6AWRR1017 pKa = 11.84TADD1020 pKa = 3.62YY1021 pKa = 11.0LVLVNSGNKK1030 pKa = 9.73LVMRR1034 pKa = 11.84ILL1036 pKa = 3.73
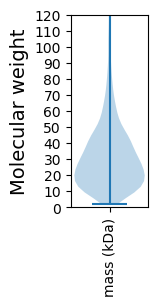
Molecular weight: 108.47 kDa
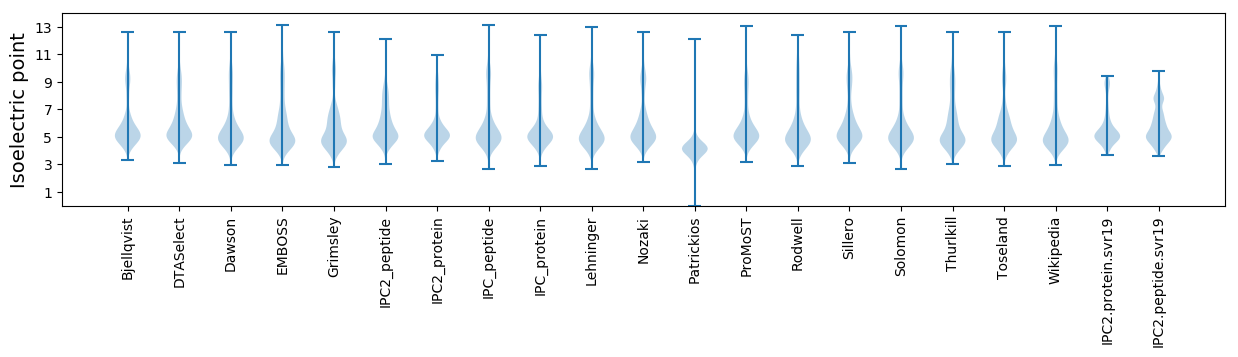
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A483CPG0|A0A483CPG0_9EURY NMD protein affecting ribosome stability and mRNA decay OS=Methanofollis sp. FWC-SCC2 OX=2052832 GN=CUJ86_04500 PE=4 SV=1
MM1 pKa = 7.65SKK3 pKa = 8.86LTKK6 pKa = 9.79GRR8 pKa = 11.84KK9 pKa = 8.52IRR11 pKa = 11.84LAKK14 pKa = 10.47ACMQNRR20 pKa = 11.84RR21 pKa = 11.84VPAWVMIRR29 pKa = 11.84TKK31 pKa = 10.5RR32 pKa = 11.84KK33 pKa = 8.78VASHH37 pKa = 6.72PKK39 pKa = 8.27RR40 pKa = 11.84RR41 pKa = 11.84MWRR44 pKa = 11.84RR45 pKa = 11.84STLKK49 pKa = 10.48VV50 pKa = 3.07
MM1 pKa = 7.65SKK3 pKa = 8.86LTKK6 pKa = 9.79GRR8 pKa = 11.84KK9 pKa = 8.52IRR11 pKa = 11.84LAKK14 pKa = 10.47ACMQNRR20 pKa = 11.84RR21 pKa = 11.84VPAWVMIRR29 pKa = 11.84TKK31 pKa = 10.5RR32 pKa = 11.84KK33 pKa = 8.78VASHH37 pKa = 6.72PKK39 pKa = 8.27RR40 pKa = 11.84RR41 pKa = 11.84MWRR44 pKa = 11.84RR45 pKa = 11.84STLKK49 pKa = 10.48VV50 pKa = 3.07
Molecular weight: 6.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
690178 |
18 |
3487 |
305.8 |
33.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.824 ± 0.075 | 1.436 ± 0.03 |
5.811 ± 0.043 | 6.977 ± 0.06 |
3.648 ± 0.035 | 8.793 ± 0.06 |
2.016 ± 0.023 | 6.748 ± 0.063 |
3.094 ± 0.044 | 9.091 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.575 ± 0.028 | 2.785 ± 0.049 |
4.822 ± 0.035 | 2.429 ± 0.027 |
6.649 ± 0.081 | 5.605 ± 0.046 |
5.816 ± 0.084 | 7.938 ± 0.05 |
1.023 ± 0.022 | 2.922 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
