
Wuhan house centipede virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KF14|A0A1L3KF14_9VIRU Capsid protein OS=Wuhan house centipede virus 5 OX=1923709 PE=3 SV=1
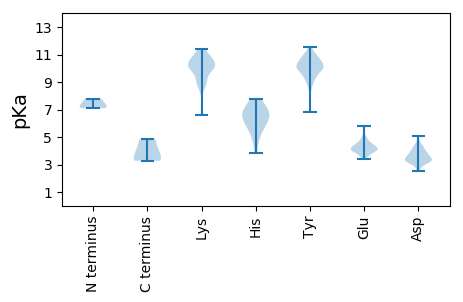
MM1 pKa = 7.1QVFGVPWGFEE11 pKa = 3.86RR12 pKa = 11.84EE13 pKa = 3.78RR14 pKa = 11.84GYY16 pKa = 10.31PRR18 pKa = 11.84MPATTPPSFNTRR30 pKa = 11.84VQEE33 pKa = 3.81AFTRR37 pKa = 11.84IKK39 pKa = 10.97NEE41 pKa = 3.45VPIISRR47 pKa = 11.84YY48 pKa = 9.9LEE50 pKa = 4.01EE51 pKa = 4.0STNYY55 pKa = 9.28VWPSRR60 pKa = 11.84QIASEE65 pKa = 4.04LKK67 pKa = 9.06SLQVHH72 pKa = 4.98SEE74 pKa = 4.28RR75 pKa = 11.84YY76 pKa = 9.2FKK78 pKa = 11.18LSTNAMHH85 pKa = 7.52PDD87 pKa = 3.66KK88 pKa = 10.69DD89 pKa = 3.91LKK91 pKa = 11.13SAILYY96 pKa = 9.35HH97 pKa = 6.78AEE99 pKa = 3.74SSYY102 pKa = 11.53GKK104 pKa = 10.4ARR106 pKa = 11.84WTLPEE111 pKa = 5.04DD112 pKa = 2.89WWSWRR117 pKa = 11.84AFKK120 pKa = 10.58EE121 pKa = 4.02AVRR124 pKa = 11.84NLDD127 pKa = 3.5MTSSPGIPYY136 pKa = 9.59MKK138 pKa = 10.05EE139 pKa = 3.41APTNGKK145 pKa = 6.64WLKK148 pKa = 9.81WDD150 pKa = 5.05GILCDD155 pKa = 3.67SLQLEE160 pKa = 4.68RR161 pKa = 11.84LWHH164 pKa = 5.98DD165 pKa = 3.46TQKK168 pKa = 11.01VIEE171 pKa = 4.51PEE173 pKa = 3.56FDD175 pKa = 3.09LLLRR179 pKa = 11.84VFIKK183 pKa = 10.43QEE185 pKa = 3.56PHH187 pKa = 6.61KK188 pKa = 10.45KK189 pKa = 9.97RR190 pKa = 11.84KK191 pKa = 9.59ADD193 pKa = 3.47EE194 pKa = 4.32DD195 pKa = 3.26RR196 pKa = 11.84WRR198 pKa = 11.84LIMAAPLSVQVAWQMCFRR216 pKa = 11.84YY217 pKa = 9.86MNDD220 pKa = 3.3LEE222 pKa = 4.26IEE224 pKa = 4.14KK225 pKa = 10.56AYY227 pKa = 10.32DD228 pKa = 3.47LPSQQGIILVSGGWKK243 pKa = 10.06VYY245 pKa = 8.55SQQWKK250 pKa = 9.07EE251 pKa = 3.84QGQTCGMDD259 pKa = 3.17KK260 pKa = 10.95SAWDD264 pKa = 3.21WTAPRR269 pKa = 11.84WALALDD275 pKa = 3.48LEE277 pKa = 4.7FRR279 pKa = 11.84KK280 pKa = 10.16RR281 pKa = 11.84LCNKK285 pKa = 9.26GMNIAWEE292 pKa = 4.82HH293 pKa = 5.61ISEE296 pKa = 4.17TLYY299 pKa = 11.08RR300 pKa = 11.84QMFDD304 pKa = 3.08NPILVLSDD312 pKa = 3.03GTAYY316 pKa = 10.31RR317 pKa = 11.84QVVPGIMKK325 pKa = 9.98SGCVNTISTNSHH337 pKa = 5.2CQVFIHH343 pKa = 6.58IAVALTYY350 pKa = 9.88KK351 pKa = 10.32IPLKK355 pKa = 10.14PLPRR359 pKa = 11.84CVGDD363 pKa = 4.28DD364 pKa = 3.7TLCTIGQSPGGIISYY379 pKa = 9.92YY380 pKa = 10.84DD381 pKa = 3.29SFGVILKK388 pKa = 10.32SISDD392 pKa = 3.51TLEE395 pKa = 3.62FVGRR399 pKa = 11.84EE400 pKa = 4.04FTDD403 pKa = 3.31EE404 pKa = 4.53GPIPMYY410 pKa = 10.14IGKK413 pKa = 8.81HH414 pKa = 3.84VVKK417 pKa = 9.79AQHH420 pKa = 6.27VRR422 pKa = 11.84DD423 pKa = 4.39EE424 pKa = 4.49DD425 pKa = 3.67LSMYY429 pKa = 9.43LDD431 pKa = 3.5SMARR435 pKa = 11.84EE436 pKa = 4.39YY437 pKa = 10.97CHH439 pKa = 7.09SPEE442 pKa = 4.01YY443 pKa = 10.68FEE445 pKa = 4.05VWKK448 pKa = 10.79RR449 pKa = 11.84LADD452 pKa = 3.77EE453 pKa = 5.19LEE455 pKa = 4.4CSLPLSRR462 pKa = 11.84EE463 pKa = 4.13AYY465 pKa = 10.11LYY467 pKa = 10.08WYY469 pKa = 8.87DD470 pKa = 4.02FPEE473 pKa = 4.48DD474 pKa = 3.28
MM1 pKa = 7.1QVFGVPWGFEE11 pKa = 3.86RR12 pKa = 11.84EE13 pKa = 3.78RR14 pKa = 11.84GYY16 pKa = 10.31PRR18 pKa = 11.84MPATTPPSFNTRR30 pKa = 11.84VQEE33 pKa = 3.81AFTRR37 pKa = 11.84IKK39 pKa = 10.97NEE41 pKa = 3.45VPIISRR47 pKa = 11.84YY48 pKa = 9.9LEE50 pKa = 4.01EE51 pKa = 4.0STNYY55 pKa = 9.28VWPSRR60 pKa = 11.84QIASEE65 pKa = 4.04LKK67 pKa = 9.06SLQVHH72 pKa = 4.98SEE74 pKa = 4.28RR75 pKa = 11.84YY76 pKa = 9.2FKK78 pKa = 11.18LSTNAMHH85 pKa = 7.52PDD87 pKa = 3.66KK88 pKa = 10.69DD89 pKa = 3.91LKK91 pKa = 11.13SAILYY96 pKa = 9.35HH97 pKa = 6.78AEE99 pKa = 3.74SSYY102 pKa = 11.53GKK104 pKa = 10.4ARR106 pKa = 11.84WTLPEE111 pKa = 5.04DD112 pKa = 2.89WWSWRR117 pKa = 11.84AFKK120 pKa = 10.58EE121 pKa = 4.02AVRR124 pKa = 11.84NLDD127 pKa = 3.5MTSSPGIPYY136 pKa = 9.59MKK138 pKa = 10.05EE139 pKa = 3.41APTNGKK145 pKa = 6.64WLKK148 pKa = 9.81WDD150 pKa = 5.05GILCDD155 pKa = 3.67SLQLEE160 pKa = 4.68RR161 pKa = 11.84LWHH164 pKa = 5.98DD165 pKa = 3.46TQKK168 pKa = 11.01VIEE171 pKa = 4.51PEE173 pKa = 3.56FDD175 pKa = 3.09LLLRR179 pKa = 11.84VFIKK183 pKa = 10.43QEE185 pKa = 3.56PHH187 pKa = 6.61KK188 pKa = 10.45KK189 pKa = 9.97RR190 pKa = 11.84KK191 pKa = 9.59ADD193 pKa = 3.47EE194 pKa = 4.32DD195 pKa = 3.26RR196 pKa = 11.84WRR198 pKa = 11.84LIMAAPLSVQVAWQMCFRR216 pKa = 11.84YY217 pKa = 9.86MNDD220 pKa = 3.3LEE222 pKa = 4.26IEE224 pKa = 4.14KK225 pKa = 10.56AYY227 pKa = 10.32DD228 pKa = 3.47LPSQQGIILVSGGWKK243 pKa = 10.06VYY245 pKa = 8.55SQQWKK250 pKa = 9.07EE251 pKa = 3.84QGQTCGMDD259 pKa = 3.17KK260 pKa = 10.95SAWDD264 pKa = 3.21WTAPRR269 pKa = 11.84WALALDD275 pKa = 3.48LEE277 pKa = 4.7FRR279 pKa = 11.84KK280 pKa = 10.16RR281 pKa = 11.84LCNKK285 pKa = 9.26GMNIAWEE292 pKa = 4.82HH293 pKa = 5.61ISEE296 pKa = 4.17TLYY299 pKa = 11.08RR300 pKa = 11.84QMFDD304 pKa = 3.08NPILVLSDD312 pKa = 3.03GTAYY316 pKa = 10.31RR317 pKa = 11.84QVVPGIMKK325 pKa = 9.98SGCVNTISTNSHH337 pKa = 5.2CQVFIHH343 pKa = 6.58IAVALTYY350 pKa = 9.88KK351 pKa = 10.32IPLKK355 pKa = 10.14PLPRR359 pKa = 11.84CVGDD363 pKa = 4.28DD364 pKa = 3.7TLCTIGQSPGGIISYY379 pKa = 9.92YY380 pKa = 10.84DD381 pKa = 3.29SFGVILKK388 pKa = 10.32SISDD392 pKa = 3.51TLEE395 pKa = 3.62FVGRR399 pKa = 11.84EE400 pKa = 4.04FTDD403 pKa = 3.31EE404 pKa = 4.53GPIPMYY410 pKa = 10.14IGKK413 pKa = 8.81HH414 pKa = 3.84VVKK417 pKa = 9.79AQHH420 pKa = 6.27VRR422 pKa = 11.84DD423 pKa = 4.39EE424 pKa = 4.49DD425 pKa = 3.67LSMYY429 pKa = 9.43LDD431 pKa = 3.5SMARR435 pKa = 11.84EE436 pKa = 4.39YY437 pKa = 10.97CHH439 pKa = 7.09SPEE442 pKa = 4.01YY443 pKa = 10.68FEE445 pKa = 4.05VWKK448 pKa = 10.79RR449 pKa = 11.84LADD452 pKa = 3.77EE453 pKa = 5.19LEE455 pKa = 4.4CSLPLSRR462 pKa = 11.84EE463 pKa = 4.13AYY465 pKa = 10.11LYY467 pKa = 10.08WYY469 pKa = 8.87DD470 pKa = 4.02FPEE473 pKa = 4.48DD474 pKa = 3.28
Molecular weight: 55.23 kDa
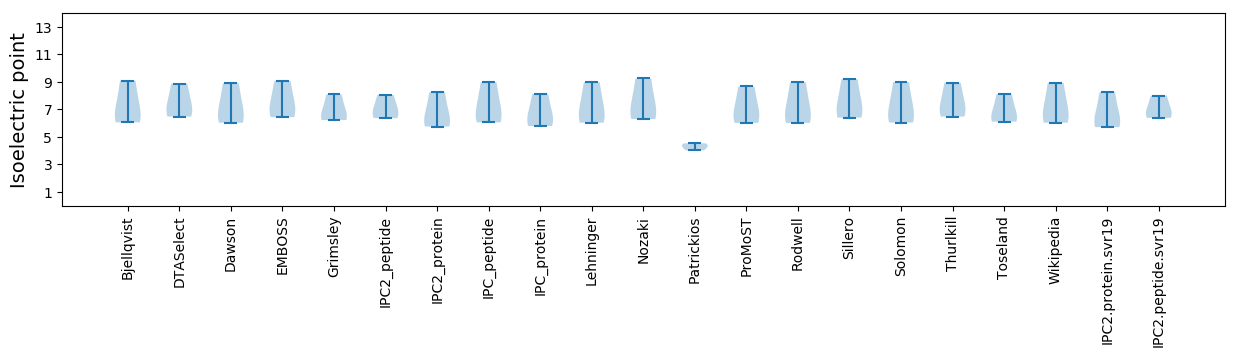
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF82|A0A1L3KF82_9VIRU Uncharacterized protein OS=Wuhan house centipede virus 5 OX=1923709 PE=4 SV=1
MM1 pKa = 7.79ASYY4 pKa = 10.6CQFSLARR11 pKa = 11.84VLDD14 pKa = 3.93MLQAPVLAPEE24 pKa = 4.08VTGNKK29 pKa = 8.86WGTLFSGLGKK39 pKa = 9.34GTLAFVNGTRR49 pKa = 11.84FGMTCARR56 pKa = 11.84NTYY59 pKa = 9.35EE60 pKa = 5.78ALPDD64 pKa = 3.34WARR67 pKa = 11.84QIILCGGVSAASIYY81 pKa = 10.3AYY83 pKa = 8.63QRR85 pKa = 11.84RR86 pKa = 11.84EE87 pKa = 3.92RR88 pKa = 11.84IKK90 pKa = 11.08DD91 pKa = 3.8RR92 pKa = 11.84IALLRR97 pKa = 11.84TMCKK101 pKa = 9.93RR102 pKa = 11.84KK103 pKa = 9.6IRR105 pKa = 11.84TIVPTPSSKK114 pKa = 10.96VYY116 pKa = 9.54MEE118 pKa = 4.21SVRR121 pKa = 11.84KK122 pKa = 9.92GSEE125 pKa = 3.89EE126 pKa = 4.42RR127 pKa = 11.84PMTSCAAQCTIGRR140 pKa = 11.84IDD142 pKa = 3.34VAGDD146 pKa = 3.51FIVIGNALRR155 pKa = 11.84FDD157 pKa = 4.45EE158 pKa = 4.92DD159 pKa = 3.77WLVGPDD165 pKa = 3.3HH166 pKa = 6.91VLAEE170 pKa = 4.24DD171 pKa = 3.23AVYY174 pKa = 10.79AKK176 pKa = 10.41GRR178 pKa = 11.84QSKK181 pKa = 10.25VSLKK185 pKa = 10.14GFEE188 pKa = 4.1RR189 pKa = 11.84VKK191 pKa = 10.84LATDD195 pKa = 3.83LVAIRR200 pKa = 11.84LKK202 pKa = 10.72SDD204 pKa = 2.53DD205 pKa = 3.24WARR208 pKa = 11.84IGVAVLKK215 pKa = 10.28LGHH218 pKa = 5.94VEE220 pKa = 3.73GSEE223 pKa = 4.28FVSICGPAGTGTCGSMSLDD242 pKa = 3.16HH243 pKa = 7.04EE244 pKa = 4.51RR245 pKa = 11.84FGWTVYY251 pKa = 10.57EE252 pKa = 4.43GTTMPGYY259 pKa = 10.57SGAAYY264 pKa = 10.02QKK266 pKa = 11.03GPMAVSMHH274 pKa = 4.52VHH276 pKa = 6.02GGVVNAGYY284 pKa = 10.11AAGYY288 pKa = 9.37IWVLLKK294 pKa = 10.3IHH296 pKa = 6.23MKK298 pKa = 9.91MYY300 pKa = 10.95DD301 pKa = 4.0EE302 pKa = 4.73DD303 pKa = 4.21TEE305 pKa = 4.76SYY307 pKa = 10.72LRR309 pKa = 11.84EE310 pKa = 3.6QHH312 pKa = 6.42RR313 pKa = 11.84RR314 pKa = 11.84GTITRR319 pKa = 11.84FRR321 pKa = 11.84SSPWGGDD328 pKa = 3.18EE329 pKa = 5.62VIFEE333 pKa = 4.13INGRR337 pKa = 11.84FEE339 pKa = 3.74FHH341 pKa = 7.54RR342 pKa = 11.84EE343 pKa = 3.52RR344 pKa = 11.84NLAAHH349 pKa = 6.91FGEE352 pKa = 4.22NWEE355 pKa = 4.17QVLRR359 pKa = 11.84PRR361 pKa = 11.84GGRR364 pKa = 11.84QYY366 pKa = 10.88SRR368 pKa = 11.84HH369 pKa = 5.46YY370 pKa = 10.03RR371 pKa = 11.84DD372 pKa = 4.17EE373 pKa = 4.25VPEE376 pKa = 4.14GANLVKK382 pKa = 10.7AKK384 pKa = 10.35EE385 pKa = 4.12EE386 pKa = 4.13PEE388 pKa = 3.77EE389 pKa = 4.36SGEE392 pKa = 4.46CKK394 pKa = 10.37CSEE397 pKa = 4.33CLGASNANVVTQGCQQQRR415 pKa = 11.84LLALTQEE422 pKa = 4.1YY423 pKa = 10.01RR424 pKa = 11.84KK425 pKa = 10.12LSRR428 pKa = 11.84ASRR431 pKa = 11.84TKK433 pKa = 9.93FRR435 pKa = 11.84SSLGILKK442 pKa = 8.72NQPITSGLQDD452 pKa = 3.9KK453 pKa = 11.02SPASS457 pKa = 3.53
MM1 pKa = 7.79ASYY4 pKa = 10.6CQFSLARR11 pKa = 11.84VLDD14 pKa = 3.93MLQAPVLAPEE24 pKa = 4.08VTGNKK29 pKa = 8.86WGTLFSGLGKK39 pKa = 9.34GTLAFVNGTRR49 pKa = 11.84FGMTCARR56 pKa = 11.84NTYY59 pKa = 9.35EE60 pKa = 5.78ALPDD64 pKa = 3.34WARR67 pKa = 11.84QIILCGGVSAASIYY81 pKa = 10.3AYY83 pKa = 8.63QRR85 pKa = 11.84RR86 pKa = 11.84EE87 pKa = 3.92RR88 pKa = 11.84IKK90 pKa = 11.08DD91 pKa = 3.8RR92 pKa = 11.84IALLRR97 pKa = 11.84TMCKK101 pKa = 9.93RR102 pKa = 11.84KK103 pKa = 9.6IRR105 pKa = 11.84TIVPTPSSKK114 pKa = 10.96VYY116 pKa = 9.54MEE118 pKa = 4.21SVRR121 pKa = 11.84KK122 pKa = 9.92GSEE125 pKa = 3.89EE126 pKa = 4.42RR127 pKa = 11.84PMTSCAAQCTIGRR140 pKa = 11.84IDD142 pKa = 3.34VAGDD146 pKa = 3.51FIVIGNALRR155 pKa = 11.84FDD157 pKa = 4.45EE158 pKa = 4.92DD159 pKa = 3.77WLVGPDD165 pKa = 3.3HH166 pKa = 6.91VLAEE170 pKa = 4.24DD171 pKa = 3.23AVYY174 pKa = 10.79AKK176 pKa = 10.41GRR178 pKa = 11.84QSKK181 pKa = 10.25VSLKK185 pKa = 10.14GFEE188 pKa = 4.1RR189 pKa = 11.84VKK191 pKa = 10.84LATDD195 pKa = 3.83LVAIRR200 pKa = 11.84LKK202 pKa = 10.72SDD204 pKa = 2.53DD205 pKa = 3.24WARR208 pKa = 11.84IGVAVLKK215 pKa = 10.28LGHH218 pKa = 5.94VEE220 pKa = 3.73GSEE223 pKa = 4.28FVSICGPAGTGTCGSMSLDD242 pKa = 3.16HH243 pKa = 7.04EE244 pKa = 4.51RR245 pKa = 11.84FGWTVYY251 pKa = 10.57EE252 pKa = 4.43GTTMPGYY259 pKa = 10.57SGAAYY264 pKa = 10.02QKK266 pKa = 11.03GPMAVSMHH274 pKa = 4.52VHH276 pKa = 6.02GGVVNAGYY284 pKa = 10.11AAGYY288 pKa = 9.37IWVLLKK294 pKa = 10.3IHH296 pKa = 6.23MKK298 pKa = 9.91MYY300 pKa = 10.95DD301 pKa = 4.0EE302 pKa = 4.73DD303 pKa = 4.21TEE305 pKa = 4.76SYY307 pKa = 10.72LRR309 pKa = 11.84EE310 pKa = 3.6QHH312 pKa = 6.42RR313 pKa = 11.84RR314 pKa = 11.84GTITRR319 pKa = 11.84FRR321 pKa = 11.84SSPWGGDD328 pKa = 3.18EE329 pKa = 5.62VIFEE333 pKa = 4.13INGRR337 pKa = 11.84FEE339 pKa = 3.74FHH341 pKa = 7.54RR342 pKa = 11.84EE343 pKa = 3.52RR344 pKa = 11.84NLAAHH349 pKa = 6.91FGEE352 pKa = 4.22NWEE355 pKa = 4.17QVLRR359 pKa = 11.84PRR361 pKa = 11.84GGRR364 pKa = 11.84QYY366 pKa = 10.88SRR368 pKa = 11.84HH369 pKa = 5.46YY370 pKa = 10.03RR371 pKa = 11.84DD372 pKa = 4.17EE373 pKa = 4.25VPEE376 pKa = 4.14GANLVKK382 pKa = 10.7AKK384 pKa = 10.35EE385 pKa = 4.12EE386 pKa = 4.13PEE388 pKa = 3.77EE389 pKa = 4.36SGEE392 pKa = 4.46CKK394 pKa = 10.37CSEE397 pKa = 4.33CLGASNANVVTQGCQQQRR415 pKa = 11.84LLALTQEE422 pKa = 4.1YY423 pKa = 10.01RR424 pKa = 11.84KK425 pKa = 10.12LSRR428 pKa = 11.84ASRR431 pKa = 11.84TKK433 pKa = 9.93FRR435 pKa = 11.84SSLGILKK442 pKa = 8.72NQPITSGLQDD452 pKa = 3.9KK453 pKa = 11.02SPASS457 pKa = 3.53
Molecular weight: 50.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1425 |
457 |
494 |
475.0 |
53.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.579 ± 0.865 | 2.035 ± 0.293 |
5.123 ± 0.494 | 6.737 ± 0.242 |
3.789 ± 0.301 | 7.158 ± 1.181 |
1.965 ± 0.255 | 5.193 ± 0.339 |
5.193 ± 0.347 | 7.649 ± 0.531 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.386 ± 0.476 | 3.298 ± 0.62 |
5.053 ± 0.627 | 3.86 ± 0.168 |
7.018 ± 0.594 | 7.088 ± 0.194 |
5.474 ± 0.609 | 6.807 ± 0.659 |
2.667 ± 0.619 | 3.86 ± 0.361 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
