
Leptobacterium flavescens
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Leptobacterium
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
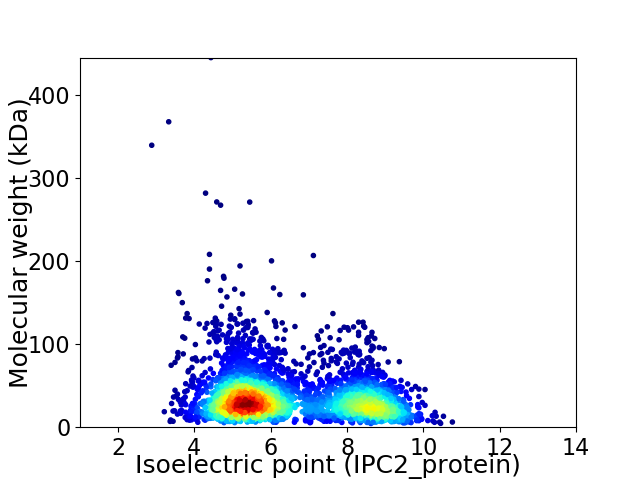
Virtual 2D-PAGE plot for 3760 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P0UI54|A0A6P0UI54_9FLAO Uncharacterized protein OS=Leptobacterium flavescens OX=472055 GN=GWK08_06195 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.34KK3 pKa = 10.37LYY5 pKa = 10.52FLICYY10 pKa = 9.62LLFGAYY16 pKa = 8.52TYY18 pKa = 11.44AQITVDD24 pKa = 3.81DD25 pKa = 4.47TSFSAQQLVEE35 pKa = 4.19EE36 pKa = 4.42VLINSPCATTSNWTASTGIIPAQGVDD62 pKa = 2.97INGIGFFEE70 pKa = 5.02RR71 pKa = 11.84GTSNFPFEE79 pKa = 4.5RR80 pKa = 11.84GVVLSSGRR88 pKa = 11.84ASLAGARR95 pKa = 11.84NPVDD99 pKa = 3.91PNLNPQSEE107 pKa = 4.77GSAPTDD113 pKa = 3.11TGGAPLWRR121 pKa = 11.84GDD123 pKa = 3.55NDD125 pKa = 3.79LEE127 pKa = 4.73NIVGLPLGNTFNASSLSFDD146 pKa = 4.28FVSQANQISFNFIFASEE163 pKa = 4.52EE164 pKa = 3.85YY165 pKa = 9.85NQNFEE170 pKa = 4.7CQFSDD175 pKa = 3.04AFAFILTDD183 pKa = 3.88SSGNSQNLAVVPNTTIPITTMTIRR207 pKa = 11.84RR208 pKa = 11.84RR209 pKa = 11.84VVPTAPGSTGCPAQNPQFFEE229 pKa = 4.52RR230 pKa = 11.84YY231 pKa = 9.51NFGPTAASSAIAFNGQTVALTASGTVVPGEE261 pKa = 4.21TYY263 pKa = 9.96TIKK266 pKa = 10.81LVVADD271 pKa = 4.27QSDD274 pKa = 3.95GALDD278 pKa = 3.4SAVFLEE284 pKa = 4.64AGSFNLGGSLGDD296 pKa = 3.96DD297 pKa = 3.12RR298 pKa = 11.84TVASGNPGCDD308 pKa = 3.11GEE310 pKa = 5.98AITLDD315 pKa = 3.47ATITGATYY323 pKa = 10.41SWFILNEE330 pKa = 4.03TTNVFEE336 pKa = 5.11PLPDD340 pKa = 3.78DD341 pKa = 3.61TNPAVNEE348 pKa = 3.99TDD350 pKa = 3.25PVINVTTSGTYY361 pKa = 9.58KK362 pKa = 10.59VEE364 pKa = 4.05GEE366 pKa = 4.54LIGTCSFEE374 pKa = 4.45DD375 pKa = 3.63EE376 pKa = 4.46VVIEE380 pKa = 5.78FITPPQIEE388 pKa = 4.82NSPQDD393 pKa = 4.94LEE395 pKa = 5.11ACDD398 pKa = 4.34EE399 pKa = 4.43GNNDD403 pKa = 3.18NAIFDD408 pKa = 4.42LSVNTPVVLGSQDD421 pKa = 3.45LNQFVVSYY429 pKa = 10.76HH430 pKa = 5.78LTQADD435 pKa = 3.54ADD437 pKa = 4.42SNSGGIDD444 pKa = 3.01PAAFSNTQNPQQIFIRR460 pKa = 11.84VADD463 pKa = 3.81NTQSCFVTANFTVTAVPLSIQALDD487 pKa = 4.84DD488 pKa = 5.32LEE490 pKa = 4.82QCDD493 pKa = 4.15PDD495 pKa = 4.15GVASFDD501 pKa = 3.79LSDD504 pKa = 3.56RR505 pKa = 11.84TSDD508 pKa = 4.19VIGTNDD514 pKa = 3.53PSRR517 pKa = 11.84VTVTYY522 pKa = 10.71HH523 pKa = 6.06LTQADD528 pKa = 4.24ADD530 pKa = 4.25TANAPLPDD538 pKa = 3.62NGYY541 pKa = 11.03DD542 pKa = 3.15NTAPVQQIFVRR553 pKa = 11.84LQDD556 pKa = 3.65NANPNCFATASFNIIVNIPPVINQVDD582 pKa = 4.22DD583 pKa = 4.76IEE585 pKa = 5.07EE586 pKa = 4.42CDD588 pKa = 3.77NDD590 pKa = 5.33LDD592 pKa = 5.63GDD594 pKa = 3.98DD595 pKa = 3.79TNGFVEE601 pKa = 5.05FDD603 pKa = 3.34LSLRR607 pKa = 11.84DD608 pKa = 3.27TDD610 pKa = 5.8LINGQANVVIDD621 pKa = 4.25SYY623 pKa = 11.38HH624 pKa = 6.72SSRR627 pKa = 11.84ADD629 pKa = 3.25AEE631 pKa = 4.76SGNSPLPAIYY641 pKa = 10.35TNTTANRR648 pKa = 11.84EE649 pKa = 4.34TIFVRR654 pKa = 11.84LRR656 pKa = 11.84NTASDD661 pKa = 4.16CVSVADD667 pKa = 4.48FDD669 pKa = 5.65LVVNPLPDD677 pKa = 3.46VTNVNLLQCDD687 pKa = 3.19VDD689 pKa = 4.1GNPDD693 pKa = 2.94GFTIYY698 pKa = 10.97NLDD701 pKa = 3.75EE702 pKa = 4.67ANIDD706 pKa = 3.86IISGGNTADD715 pKa = 3.38FTFTYY720 pKa = 10.25FLSLADD726 pKa = 3.85AQSGNNPQAPAPFTNTANPQTLFVSVVNNTTSCSRR761 pKa = 11.84IVEE764 pKa = 4.5VILDD768 pKa = 3.54VTATNVSNASLTLCDD783 pKa = 4.49DD784 pKa = 4.25DD785 pKa = 6.33RR786 pKa = 11.84DD787 pKa = 3.91GSVAFDD793 pKa = 3.71LTLADD798 pKa = 3.88AQVLTSAPPGLDD810 pKa = 2.81VAYY813 pKa = 10.64YY814 pKa = 8.71ATLQDD819 pKa = 3.98AQLEE823 pKa = 4.51QNRR826 pKa = 11.84LPDD829 pKa = 4.21LYY831 pKa = 11.03SNSSNPQTVFARR843 pKa = 11.84VEE845 pKa = 3.82NANACFGISSLTLNVNEE862 pKa = 5.48LPPQNPLSDD871 pKa = 4.39FVVCDD876 pKa = 4.46DD877 pKa = 5.42DD878 pKa = 4.76DD879 pKa = 4.02TFTFDD884 pKa = 5.86LSTKK888 pKa = 9.53DD889 pKa = 3.51AEE891 pKa = 4.83VFGSANPADD900 pKa = 4.02FTVSYY905 pKa = 10.36HH906 pKa = 7.18SNDD909 pKa = 2.9ADD911 pKa = 5.0AIADD915 pKa = 4.14TNPLTTNYY923 pKa = 9.88TNISNPQTVVARR935 pKa = 11.84LEE937 pKa = 4.1NNITGCFVTTAFRR950 pKa = 11.84LSVNINPTVNQPLPLEE966 pKa = 4.4ICDD969 pKa = 4.41DD970 pKa = 4.84DD971 pKa = 4.57IVDD974 pKa = 4.37GFAAFDD980 pKa = 3.75LTTKK984 pKa = 10.33NAEE987 pKa = 3.62ITGNQNGVEE996 pKa = 3.89VSYY999 pKa = 11.49YY1000 pKa = 9.51LTQNDD1005 pKa = 3.98ADD1007 pKa = 3.87MGINPLPDD1015 pKa = 3.17TYY1017 pKa = 11.5TNISNPQIVFVRR1029 pKa = 11.84VQNTTTSCFRR1039 pKa = 11.84TDD1041 pKa = 3.02NTTLEE1046 pKa = 5.1LIVQQGAVANRR1057 pKa = 11.84PQPLQTCDD1065 pKa = 3.44EE1066 pKa = 4.56LGEE1069 pKa = 4.27NDD1071 pKa = 5.56GIAQFDD1077 pKa = 3.93LTALDD1082 pKa = 3.6NEE1084 pKa = 4.32IRR1086 pKa = 11.84GNQDD1090 pKa = 2.59PSQYY1094 pKa = 9.77RR1095 pKa = 11.84VSYY1098 pKa = 10.6HH1099 pKa = 5.14LTEE1102 pKa = 5.03SDD1104 pKa = 3.87AEE1106 pKa = 4.31AGSNEE1111 pKa = 3.83INGAYY1116 pKa = 9.45TNISNPQTIYY1126 pKa = 11.3ARR1128 pKa = 11.84VTNIPIGCFNTTTAEE1143 pKa = 4.11LVVNLLPEE1151 pKa = 4.55PEE1153 pKa = 4.95LGDD1156 pKa = 3.81TVALCVDD1163 pKa = 3.77ANGTITSRR1171 pKa = 11.84PTLNSGIPASGFSFEE1186 pKa = 4.0WRR1188 pKa = 11.84LDD1190 pKa = 3.54GTLLAGEE1197 pKa = 4.5NNPDD1201 pKa = 3.58LVANTAGTYY1210 pKa = 7.62TVSVVNNSTGCRR1222 pKa = 11.84NTDD1225 pKa = 3.07EE1226 pKa = 4.41VVVRR1230 pKa = 11.84RR1231 pKa = 11.84SSPPITFSAEE1241 pKa = 3.83VVTNAFVQNNRR1252 pKa = 11.84IEE1254 pKa = 4.3VNVTGEE1260 pKa = 3.66GDD1262 pKa = 3.76YY1263 pKa = 11.05QFSLDD1268 pKa = 3.86GGSFQDD1274 pKa = 3.74DD1275 pKa = 3.84PFFEE1279 pKa = 4.4RR1280 pKa = 11.84VSPGPHH1286 pKa = 6.95TITIRR1291 pKa = 11.84DD1292 pKa = 3.44INGCGSVEE1300 pKa = 3.8VDD1302 pKa = 3.79VFIIDD1307 pKa = 3.52YY1308 pKa = 9.85PRR1310 pKa = 11.84FFTPNGDD1317 pKa = 3.66GFNDD1321 pKa = 2.97TWNIVGATRR1330 pKa = 11.84LGNPNIFIFDD1340 pKa = 3.82RR1341 pKa = 11.84YY1342 pKa = 10.62GKK1344 pKa = 9.76LLKK1347 pKa = 10.45QIEE1350 pKa = 4.22PAGNGWNGMFNGQPLPSSDD1369 pKa = 3.01YY1370 pKa = 10.33WFLINYY1376 pKa = 8.29TEE1378 pKa = 5.32DD1379 pKa = 3.84DD1380 pKa = 3.23VSKK1383 pKa = 10.68QLRR1386 pKa = 11.84GHH1388 pKa = 6.63FALKK1392 pKa = 10.31RR1393 pKa = 3.61
MM1 pKa = 7.49KK2 pKa = 10.34KK3 pKa = 10.37LYY5 pKa = 10.52FLICYY10 pKa = 9.62LLFGAYY16 pKa = 8.52TYY18 pKa = 11.44AQITVDD24 pKa = 3.81DD25 pKa = 4.47TSFSAQQLVEE35 pKa = 4.19EE36 pKa = 4.42VLINSPCATTSNWTASTGIIPAQGVDD62 pKa = 2.97INGIGFFEE70 pKa = 5.02RR71 pKa = 11.84GTSNFPFEE79 pKa = 4.5RR80 pKa = 11.84GVVLSSGRR88 pKa = 11.84ASLAGARR95 pKa = 11.84NPVDD99 pKa = 3.91PNLNPQSEE107 pKa = 4.77GSAPTDD113 pKa = 3.11TGGAPLWRR121 pKa = 11.84GDD123 pKa = 3.55NDD125 pKa = 3.79LEE127 pKa = 4.73NIVGLPLGNTFNASSLSFDD146 pKa = 4.28FVSQANQISFNFIFASEE163 pKa = 4.52EE164 pKa = 3.85YY165 pKa = 9.85NQNFEE170 pKa = 4.7CQFSDD175 pKa = 3.04AFAFILTDD183 pKa = 3.88SSGNSQNLAVVPNTTIPITTMTIRR207 pKa = 11.84RR208 pKa = 11.84RR209 pKa = 11.84VVPTAPGSTGCPAQNPQFFEE229 pKa = 4.52RR230 pKa = 11.84YY231 pKa = 9.51NFGPTAASSAIAFNGQTVALTASGTVVPGEE261 pKa = 4.21TYY263 pKa = 9.96TIKK266 pKa = 10.81LVVADD271 pKa = 4.27QSDD274 pKa = 3.95GALDD278 pKa = 3.4SAVFLEE284 pKa = 4.64AGSFNLGGSLGDD296 pKa = 3.96DD297 pKa = 3.12RR298 pKa = 11.84TVASGNPGCDD308 pKa = 3.11GEE310 pKa = 5.98AITLDD315 pKa = 3.47ATITGATYY323 pKa = 10.41SWFILNEE330 pKa = 4.03TTNVFEE336 pKa = 5.11PLPDD340 pKa = 3.78DD341 pKa = 3.61TNPAVNEE348 pKa = 3.99TDD350 pKa = 3.25PVINVTTSGTYY361 pKa = 9.58KK362 pKa = 10.59VEE364 pKa = 4.05GEE366 pKa = 4.54LIGTCSFEE374 pKa = 4.45DD375 pKa = 3.63EE376 pKa = 4.46VVIEE380 pKa = 5.78FITPPQIEE388 pKa = 4.82NSPQDD393 pKa = 4.94LEE395 pKa = 5.11ACDD398 pKa = 4.34EE399 pKa = 4.43GNNDD403 pKa = 3.18NAIFDD408 pKa = 4.42LSVNTPVVLGSQDD421 pKa = 3.45LNQFVVSYY429 pKa = 10.76HH430 pKa = 5.78LTQADD435 pKa = 3.54ADD437 pKa = 4.42SNSGGIDD444 pKa = 3.01PAAFSNTQNPQQIFIRR460 pKa = 11.84VADD463 pKa = 3.81NTQSCFVTANFTVTAVPLSIQALDD487 pKa = 4.84DD488 pKa = 5.32LEE490 pKa = 4.82QCDD493 pKa = 4.15PDD495 pKa = 4.15GVASFDD501 pKa = 3.79LSDD504 pKa = 3.56RR505 pKa = 11.84TSDD508 pKa = 4.19VIGTNDD514 pKa = 3.53PSRR517 pKa = 11.84VTVTYY522 pKa = 10.71HH523 pKa = 6.06LTQADD528 pKa = 4.24ADD530 pKa = 4.25TANAPLPDD538 pKa = 3.62NGYY541 pKa = 11.03DD542 pKa = 3.15NTAPVQQIFVRR553 pKa = 11.84LQDD556 pKa = 3.65NANPNCFATASFNIIVNIPPVINQVDD582 pKa = 4.22DD583 pKa = 4.76IEE585 pKa = 5.07EE586 pKa = 4.42CDD588 pKa = 3.77NDD590 pKa = 5.33LDD592 pKa = 5.63GDD594 pKa = 3.98DD595 pKa = 3.79TNGFVEE601 pKa = 5.05FDD603 pKa = 3.34LSLRR607 pKa = 11.84DD608 pKa = 3.27TDD610 pKa = 5.8LINGQANVVIDD621 pKa = 4.25SYY623 pKa = 11.38HH624 pKa = 6.72SSRR627 pKa = 11.84ADD629 pKa = 3.25AEE631 pKa = 4.76SGNSPLPAIYY641 pKa = 10.35TNTTANRR648 pKa = 11.84EE649 pKa = 4.34TIFVRR654 pKa = 11.84LRR656 pKa = 11.84NTASDD661 pKa = 4.16CVSVADD667 pKa = 4.48FDD669 pKa = 5.65LVVNPLPDD677 pKa = 3.46VTNVNLLQCDD687 pKa = 3.19VDD689 pKa = 4.1GNPDD693 pKa = 2.94GFTIYY698 pKa = 10.97NLDD701 pKa = 3.75EE702 pKa = 4.67ANIDD706 pKa = 3.86IISGGNTADD715 pKa = 3.38FTFTYY720 pKa = 10.25FLSLADD726 pKa = 3.85AQSGNNPQAPAPFTNTANPQTLFVSVVNNTTSCSRR761 pKa = 11.84IVEE764 pKa = 4.5VILDD768 pKa = 3.54VTATNVSNASLTLCDD783 pKa = 4.49DD784 pKa = 4.25DD785 pKa = 6.33RR786 pKa = 11.84DD787 pKa = 3.91GSVAFDD793 pKa = 3.71LTLADD798 pKa = 3.88AQVLTSAPPGLDD810 pKa = 2.81VAYY813 pKa = 10.64YY814 pKa = 8.71ATLQDD819 pKa = 3.98AQLEE823 pKa = 4.51QNRR826 pKa = 11.84LPDD829 pKa = 4.21LYY831 pKa = 11.03SNSSNPQTVFARR843 pKa = 11.84VEE845 pKa = 3.82NANACFGISSLTLNVNEE862 pKa = 5.48LPPQNPLSDD871 pKa = 4.39FVVCDD876 pKa = 4.46DD877 pKa = 5.42DD878 pKa = 4.76DD879 pKa = 4.02TFTFDD884 pKa = 5.86LSTKK888 pKa = 9.53DD889 pKa = 3.51AEE891 pKa = 4.83VFGSANPADD900 pKa = 4.02FTVSYY905 pKa = 10.36HH906 pKa = 7.18SNDD909 pKa = 2.9ADD911 pKa = 5.0AIADD915 pKa = 4.14TNPLTTNYY923 pKa = 9.88TNISNPQTVVARR935 pKa = 11.84LEE937 pKa = 4.1NNITGCFVTTAFRR950 pKa = 11.84LSVNINPTVNQPLPLEE966 pKa = 4.4ICDD969 pKa = 4.41DD970 pKa = 4.84DD971 pKa = 4.57IVDD974 pKa = 4.37GFAAFDD980 pKa = 3.75LTTKK984 pKa = 10.33NAEE987 pKa = 3.62ITGNQNGVEE996 pKa = 3.89VSYY999 pKa = 11.49YY1000 pKa = 9.51LTQNDD1005 pKa = 3.98ADD1007 pKa = 3.87MGINPLPDD1015 pKa = 3.17TYY1017 pKa = 11.5TNISNPQIVFVRR1029 pKa = 11.84VQNTTTSCFRR1039 pKa = 11.84TDD1041 pKa = 3.02NTTLEE1046 pKa = 5.1LIVQQGAVANRR1057 pKa = 11.84PQPLQTCDD1065 pKa = 3.44EE1066 pKa = 4.56LGEE1069 pKa = 4.27NDD1071 pKa = 5.56GIAQFDD1077 pKa = 3.93LTALDD1082 pKa = 3.6NEE1084 pKa = 4.32IRR1086 pKa = 11.84GNQDD1090 pKa = 2.59PSQYY1094 pKa = 9.77RR1095 pKa = 11.84VSYY1098 pKa = 10.6HH1099 pKa = 5.14LTEE1102 pKa = 5.03SDD1104 pKa = 3.87AEE1106 pKa = 4.31AGSNEE1111 pKa = 3.83INGAYY1116 pKa = 9.45TNISNPQTIYY1126 pKa = 11.3ARR1128 pKa = 11.84VTNIPIGCFNTTTAEE1143 pKa = 4.11LVVNLLPEE1151 pKa = 4.55PEE1153 pKa = 4.95LGDD1156 pKa = 3.81TVALCVDD1163 pKa = 3.77ANGTITSRR1171 pKa = 11.84PTLNSGIPASGFSFEE1186 pKa = 4.0WRR1188 pKa = 11.84LDD1190 pKa = 3.54GTLLAGEE1197 pKa = 4.5NNPDD1201 pKa = 3.58LVANTAGTYY1210 pKa = 7.62TVSVVNNSTGCRR1222 pKa = 11.84NTDD1225 pKa = 3.07EE1226 pKa = 4.41VVVRR1230 pKa = 11.84RR1231 pKa = 11.84SSPPITFSAEE1241 pKa = 3.83VVTNAFVQNNRR1252 pKa = 11.84IEE1254 pKa = 4.3VNVTGEE1260 pKa = 3.66GDD1262 pKa = 3.76YY1263 pKa = 11.05QFSLDD1268 pKa = 3.86GGSFQDD1274 pKa = 3.74DD1275 pKa = 3.84PFFEE1279 pKa = 4.4RR1280 pKa = 11.84VSPGPHH1286 pKa = 6.95TITIRR1291 pKa = 11.84DD1292 pKa = 3.44INGCGSVEE1300 pKa = 3.8VDD1302 pKa = 3.79VFIIDD1307 pKa = 3.52YY1308 pKa = 9.85PRR1310 pKa = 11.84FFTPNGDD1317 pKa = 3.66GFNDD1321 pKa = 2.97TWNIVGATRR1330 pKa = 11.84LGNPNIFIFDD1340 pKa = 3.82RR1341 pKa = 11.84YY1342 pKa = 10.62GKK1344 pKa = 9.76LLKK1347 pKa = 10.45QIEE1350 pKa = 4.22PAGNGWNGMFNGQPLPSSDD1369 pKa = 3.01YY1370 pKa = 10.33WFLINYY1376 pKa = 8.29TEE1378 pKa = 5.32DD1379 pKa = 3.84DD1380 pKa = 3.23VSKK1383 pKa = 10.68QLRR1386 pKa = 11.84GHH1388 pKa = 6.63FALKK1392 pKa = 10.31RR1393 pKa = 3.61
Molecular weight: 149.97 kDa
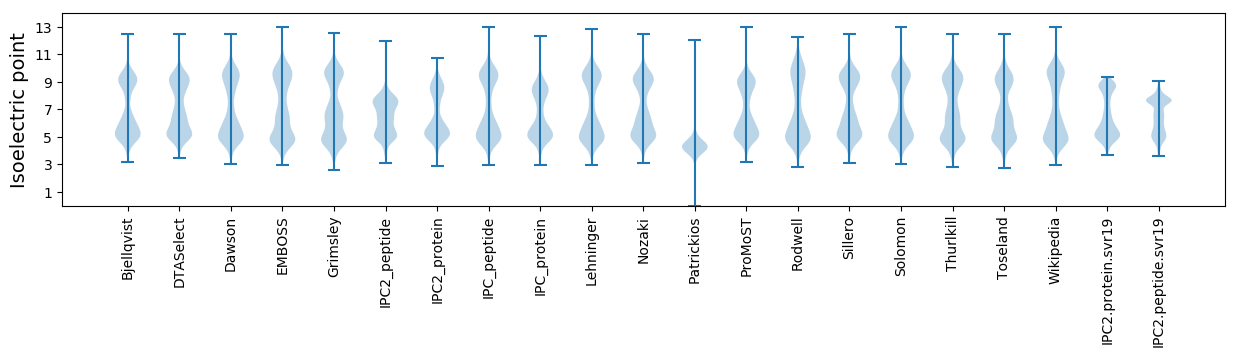
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P0UJK0|A0A6P0UJK0_9FLAO Uncharacterized protein OS=Leptobacterium flavescens OX=472055 GN=GWK08_08510 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.92KK51 pKa = 10.61KK52 pKa = 9.84
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.92KK51 pKa = 10.61KK52 pKa = 9.84
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1265630 |
38 |
4047 |
336.6 |
38.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.409 ± 0.042 | 0.71 ± 0.014 |
5.749 ± 0.044 | 7.072 ± 0.042 |
5.309 ± 0.04 | 6.686 ± 0.038 |
1.783 ± 0.018 | 7.633 ± 0.041 |
7.381 ± 0.053 | 9.419 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.104 ± 0.023 | 5.694 ± 0.04 |
3.451 ± 0.022 | 3.236 ± 0.019 |
4.152 ± 0.027 | 6.558 ± 0.031 |
5.372 ± 0.051 | 6.147 ± 0.03 |
1.094 ± 0.017 | 4.041 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
