
Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Corynebacteriaceae; Corynebacterium; Corynebacterium glutamicum
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
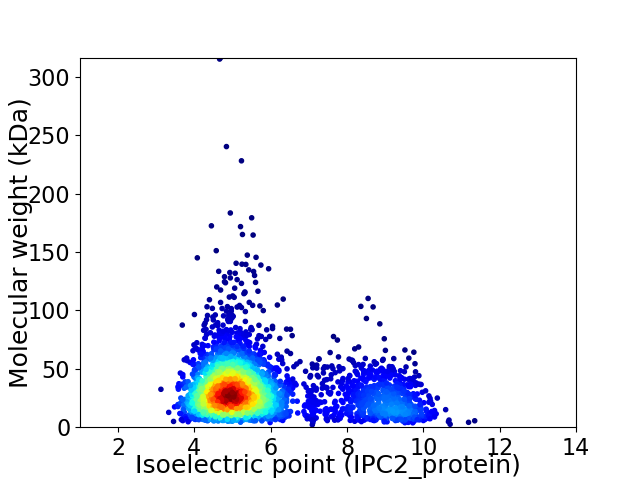
Virtual 2D-PAGE plot for 3094 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8NQT0|Q8NQT0_CORGL Uncharacterized protein OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=Cgl1344 PE=4 SV=1
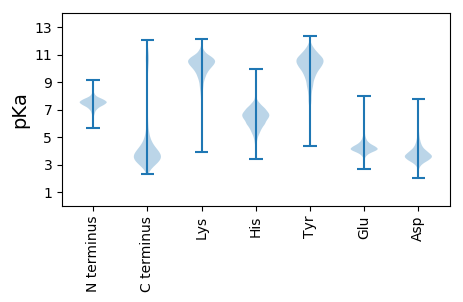
MM1 pKa = 7.74KK2 pKa = 9.92DD3 pKa = 3.48YY4 pKa = 11.05AVHH7 pKa = 5.54TRR9 pKa = 11.84GLVSLLSLIFLFVLSGCGGNATTADD34 pKa = 3.69EE35 pKa = 4.37AAEE38 pKa = 4.11SDD40 pKa = 4.13VVTSSSAPPSKK51 pKa = 10.21RR52 pKa = 11.84ALDD55 pKa = 3.35VGEE58 pKa = 4.18AVEE61 pKa = 4.26IPGVVLTVNSVTQSDD76 pKa = 3.68QLMLYY81 pKa = 10.17SEE83 pKa = 5.07GSARR87 pKa = 11.84GSEE90 pKa = 3.87PRR92 pKa = 11.84EE93 pKa = 3.79QRR95 pKa = 11.84NAASGEE101 pKa = 4.14KK102 pKa = 9.73FVSVDD107 pKa = 3.38TTVKK111 pKa = 10.65NSSSDD116 pKa = 3.36PWDD119 pKa = 3.78LSCGHH124 pKa = 6.6VLQTWLLEE132 pKa = 4.58DD133 pKa = 4.73EE134 pKa = 5.95LDD136 pKa = 4.32DD137 pKa = 4.66QQGDD141 pKa = 3.77QEE143 pKa = 5.03KK144 pKa = 10.48KK145 pKa = 10.49FSPIDD150 pKa = 3.5NLDD153 pKa = 3.78QISGNPEE160 pKa = 3.79CGVLLEE166 pKa = 4.38VGTEE170 pKa = 4.48IEE172 pKa = 4.47MTWSFTIPDD181 pKa = 5.04DD182 pKa = 4.59IEE184 pKa = 3.86ITHH187 pKa = 6.81FGFSLSDD194 pKa = 3.36STSNDD199 pKa = 2.94LAIISLGGAIEE210 pKa = 4.17TSSAITTTEE219 pKa = 3.87VIAPEE224 pKa = 4.16NDD226 pKa = 3.17TEE228 pKa = 4.35TLLEE232 pKa = 4.17ITPVDD237 pKa = 3.99CQVGLGPIVTSWSDD251 pKa = 3.27GTVGGWSQHH260 pKa = 5.58CQDD263 pKa = 3.44VHH265 pKa = 8.84DD266 pKa = 4.55EE267 pKa = 4.19VLAGEE272 pKa = 4.57VAANTPVCDD281 pKa = 4.13GVVCTYY287 pKa = 10.74PSGATMPDD295 pKa = 3.33PNAPQIPSDD304 pKa = 3.56TSGAVCDD311 pKa = 4.57EE312 pKa = 4.33NQCVYY317 pKa = 10.6PNGYY321 pKa = 8.41IARR324 pKa = 11.84IGDD327 pKa = 3.94PNVPNYY333 pKa = 9.87LKK335 pKa = 10.35PGNSPWVQGQIDD347 pKa = 4.06FQNCLDD353 pKa = 3.69SGKK356 pKa = 9.11TIEE359 pKa = 4.16QCRR362 pKa = 11.84EE363 pKa = 3.7EE364 pKa = 4.47LNN366 pKa = 3.6
MM1 pKa = 7.74KK2 pKa = 9.92DD3 pKa = 3.48YY4 pKa = 11.05AVHH7 pKa = 5.54TRR9 pKa = 11.84GLVSLLSLIFLFVLSGCGGNATTADD34 pKa = 3.69EE35 pKa = 4.37AAEE38 pKa = 4.11SDD40 pKa = 4.13VVTSSSAPPSKK51 pKa = 10.21RR52 pKa = 11.84ALDD55 pKa = 3.35VGEE58 pKa = 4.18AVEE61 pKa = 4.26IPGVVLTVNSVTQSDD76 pKa = 3.68QLMLYY81 pKa = 10.17SEE83 pKa = 5.07GSARR87 pKa = 11.84GSEE90 pKa = 3.87PRR92 pKa = 11.84EE93 pKa = 3.79QRR95 pKa = 11.84NAASGEE101 pKa = 4.14KK102 pKa = 9.73FVSVDD107 pKa = 3.38TTVKK111 pKa = 10.65NSSSDD116 pKa = 3.36PWDD119 pKa = 3.78LSCGHH124 pKa = 6.6VLQTWLLEE132 pKa = 4.58DD133 pKa = 4.73EE134 pKa = 5.95LDD136 pKa = 4.32DD137 pKa = 4.66QQGDD141 pKa = 3.77QEE143 pKa = 5.03KK144 pKa = 10.48KK145 pKa = 10.49FSPIDD150 pKa = 3.5NLDD153 pKa = 3.78QISGNPEE160 pKa = 3.79CGVLLEE166 pKa = 4.38VGTEE170 pKa = 4.48IEE172 pKa = 4.47MTWSFTIPDD181 pKa = 5.04DD182 pKa = 4.59IEE184 pKa = 3.86ITHH187 pKa = 6.81FGFSLSDD194 pKa = 3.36STSNDD199 pKa = 2.94LAIISLGGAIEE210 pKa = 4.17TSSAITTTEE219 pKa = 3.87VIAPEE224 pKa = 4.16NDD226 pKa = 3.17TEE228 pKa = 4.35TLLEE232 pKa = 4.17ITPVDD237 pKa = 3.99CQVGLGPIVTSWSDD251 pKa = 3.27GTVGGWSQHH260 pKa = 5.58CQDD263 pKa = 3.44VHH265 pKa = 8.84DD266 pKa = 4.55EE267 pKa = 4.19VLAGEE272 pKa = 4.57VAANTPVCDD281 pKa = 4.13GVVCTYY287 pKa = 10.74PSGATMPDD295 pKa = 3.33PNAPQIPSDD304 pKa = 3.56TSGAVCDD311 pKa = 4.57EE312 pKa = 4.33NQCVYY317 pKa = 10.6PNGYY321 pKa = 8.41IARR324 pKa = 11.84IGDD327 pKa = 3.94PNVPNYY333 pKa = 9.87LKK335 pKa = 10.35PGNSPWVQGQIDD347 pKa = 4.06FQNCLDD353 pKa = 3.69SGKK356 pKa = 9.11TIEE359 pKa = 4.16QCRR362 pKa = 11.84EE363 pKa = 3.7EE364 pKa = 4.47LNN366 pKa = 3.6
Molecular weight: 38.94 kDa
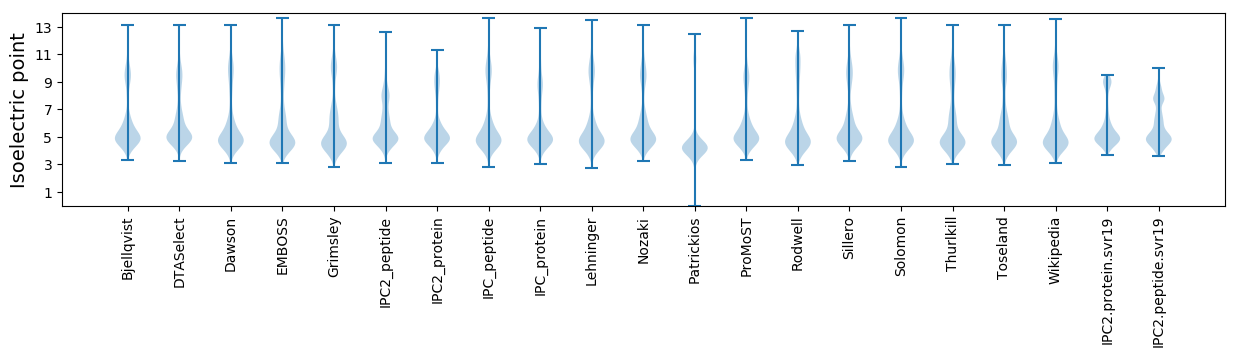
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q79VC7|Q79VC7_CORGL Permeases of the major facilitator superfamily OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=Cgl2777 PE=4 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.72GRR42 pKa = 11.84AKK44 pKa = 9.63LTAA47 pKa = 4.21
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.72GRR42 pKa = 11.84AKK44 pKa = 9.63LTAA47 pKa = 4.21
Molecular weight: 5.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
958486 |
17 |
2993 |
309.8 |
33.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.723 ± 0.049 | 0.669 ± 0.013 |
5.945 ± 0.04 | 6.354 ± 0.041 |
3.635 ± 0.029 | 8.034 ± 0.04 |
2.12 ± 0.021 | 5.775 ± 0.036 |
3.487 ± 0.032 | 9.673 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.323 ± 0.018 | 3.295 ± 0.025 |
4.842 ± 0.032 | 3.383 ± 0.029 |
5.596 ± 0.039 | 6.393 ± 0.032 |
6.172 ± 0.036 | 7.951 ± 0.039 |
1.411 ± 0.018 | 2.22 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
