
Changjiang tombus-like virus 21
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
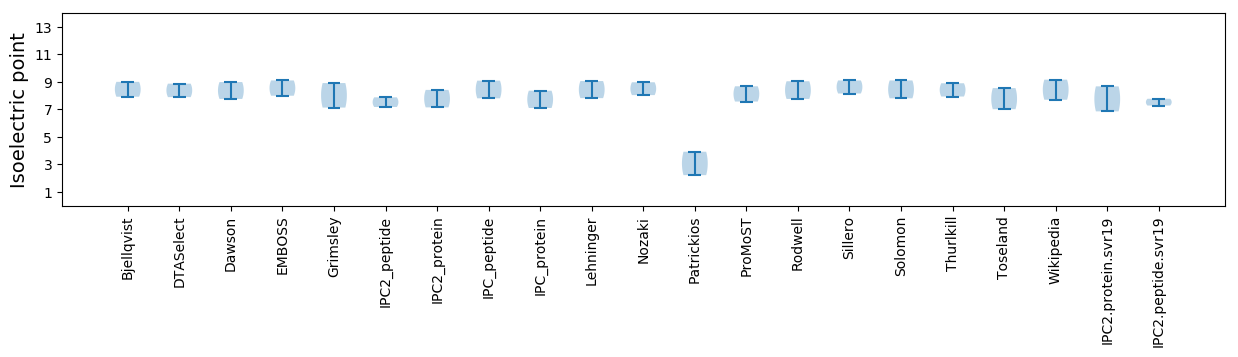
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFQ5|A0A1L3KFQ5_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 21 OX=1922815 PE=4 SV=1
MM1 pKa = 7.52EE2 pKa = 4.69NKK4 pKa = 9.92IYY6 pKa = 10.13QLPEE10 pKa = 4.32FIKK13 pKa = 10.44HH14 pKa = 4.74VPVANRR20 pKa = 11.84ADD22 pKa = 3.8YY23 pKa = 10.43IYY25 pKa = 11.13DD26 pKa = 3.56KK27 pKa = 10.87LYY29 pKa = 9.54KK30 pKa = 9.34TGSRR34 pKa = 11.84YY35 pKa = 10.2SVTDD39 pKa = 3.39HH40 pKa = 6.45SQFEE44 pKa = 4.11AHH46 pKa = 5.72FTRR49 pKa = 11.84NIMEE53 pKa = 4.9HH54 pKa = 7.33IEE56 pKa = 4.19FVVYY60 pKa = 10.16RR61 pKa = 11.84YY62 pKa = 10.23LIQNLPNKK70 pKa = 9.76EE71 pKa = 4.45DD72 pKa = 3.55YY73 pKa = 9.94EE74 pKa = 4.27QLFKK78 pKa = 11.22VLQGKK83 pKa = 7.96NKK85 pKa = 9.11CCFKK89 pKa = 10.32KK90 pKa = 10.48GHH92 pKa = 6.45YY93 pKa = 9.07IVNATRR99 pKa = 11.84MSGEE103 pKa = 4.2MNTSLGNGLTNLLLMLYY120 pKa = 10.08VLNQNGNTNVSGVVEE135 pKa = 4.31GDD137 pKa = 3.66DD138 pKa = 3.91GLFRR142 pKa = 11.84YY143 pKa = 9.67DD144 pKa = 4.92GPDD147 pKa = 3.15PTKK150 pKa = 11.51DD151 pKa = 3.03MFAKK155 pKa = 10.02MGFTVKK161 pKa = 10.9LEE163 pKa = 3.86IFDD166 pKa = 4.43NLNEE170 pKa = 4.06ASFCGLVFDD179 pKa = 4.75IDD181 pKa = 3.38ARR183 pKa = 11.84QIIINPIKK191 pKa = 10.67VLLNFGWLHH200 pKa = 5.0YY201 pKa = 9.67QYY203 pKa = 11.32KK204 pKa = 9.76DD205 pKa = 3.19AKK207 pKa = 10.16FEE209 pKa = 4.62TIMGLLRR216 pKa = 11.84MKK218 pKa = 10.39SLSLLYY224 pKa = 9.85QYY226 pKa = 9.11PSCPIVSEE234 pKa = 3.88LALYY238 pKa = 9.45GLRR241 pKa = 11.84VTSKK245 pKa = 8.4YY246 pKa = 8.93TPVLIRR252 pKa = 11.84EE253 pKa = 4.17NNYY256 pKa = 9.12KK257 pKa = 10.36RR258 pKa = 11.84KK259 pKa = 9.04FHH261 pKa = 6.63EE262 pKa = 4.38EE263 pKa = 3.68VLRR266 pKa = 11.84TPRR269 pKa = 11.84LVADD273 pKa = 4.47MINFKK278 pKa = 9.79TRR280 pKa = 11.84LVCEE284 pKa = 4.16KK285 pKa = 10.28VFKK288 pKa = 10.07ITIDD292 pKa = 3.56DD293 pKa = 3.67QLSIEE298 pKa = 4.69SYY300 pKa = 10.64LKK302 pKa = 10.39QLDD305 pKa = 4.04EE306 pKa = 5.47IKK308 pKa = 10.44PLWHH312 pKa = 6.79LSFHH316 pKa = 6.68TYY318 pKa = 8.58MNQYY322 pKa = 9.57QDD324 pKa = 4.19VYY326 pKa = 9.27LQYY329 pKa = 10.65VGEE332 pKa = 4.27YY333 pKa = 8.98AAGYY337 pKa = 7.46HH338 pKa = 4.91TVVPHH343 pKa = 6.38KK344 pKa = 9.63MVRR347 pKa = 11.84RR348 pKa = 11.84NRR350 pKa = 11.84TKK352 pKa = 9.35QQKK355 pKa = 9.22KK356 pKa = 10.23RR357 pKa = 11.84NNSNNNNNNNNNSKK371 pKa = 10.23KK372 pKa = 10.35KK373 pKa = 9.13EE374 pKa = 3.99IVVVRR379 pKa = 11.84QNNPKK384 pKa = 6.94QTKK387 pKa = 10.39SIGQQIGSALGGLIGHH403 pKa = 6.89GAHH406 pKa = 6.22MLFKK410 pKa = 10.61NITGFGDD417 pKa = 3.62YY418 pKa = 10.75NINNNTLLQGGMTPPEE434 pKa = 4.43VINTVRR440 pKa = 11.84EE441 pKa = 3.96GGFVVRR447 pKa = 11.84HH448 pKa = 5.67RR449 pKa = 11.84EE450 pKa = 4.04YY451 pKa = 10.56IGDD454 pKa = 3.9ISPSQQFSSTVYY466 pKa = 10.63SINPGNANTFPWLSQVADD484 pKa = 3.45SFEE487 pKa = 4.19QYY489 pKa = 9.7EE490 pKa = 4.08FRR492 pKa = 11.84GLVFEE497 pKa = 5.1FKK499 pKa = 9.7STSSDD504 pKa = 3.17AVLSSAASSSLGTVIMATQYY524 pKa = 11.46NSLSNPFTDD533 pKa = 3.12KK534 pKa = 10.62RR535 pKa = 11.84TMEE538 pKa = 4.08NYY540 pKa = 10.06EE541 pKa = 4.04YY542 pKa = 11.24ANSDD546 pKa = 3.6KK547 pKa = 11.03PSMSFYY553 pKa = 11.07HH554 pKa = 6.54PVEE557 pKa = 4.42CKK559 pKa = 10.36KK560 pKa = 10.76SQTSIDD566 pKa = 3.61LLYY569 pKa = 11.04VRR571 pKa = 11.84NTDD574 pKa = 3.54AVNGDD579 pKa = 3.42LRR581 pKa = 11.84LYY583 pKa = 10.94DD584 pKa = 4.34LGQFQISTTGMQNTGGVIGEE604 pKa = 4.28LWCTFEE610 pKa = 4.22IAFYY614 pKa = 10.4KK615 pKa = 10.55PKK617 pKa = 10.27LLPSGTTCLTDD628 pKa = 3.69HH629 pKa = 6.53YY630 pKa = 10.23RR631 pKa = 11.84LDD633 pKa = 5.44AITTDD638 pKa = 2.78VWFGDD643 pKa = 3.98TQTLMAGSNIGTNITTGGTRR663 pKa = 11.84IQFPATLSTGKK674 pKa = 10.32FSVVYY679 pKa = 9.73SISSITGFNTPPTISGSGLNLLPLYY704 pKa = 10.86NGGTTSTVIAAQSTLSTAIIGVTVQITDD732 pKa = 3.59PNAYY736 pKa = 10.33LDD738 pKa = 4.08FVGGAIGAAAEE749 pKa = 4.22GDD751 pKa = 3.88LFISQIALEE760 pKa = 4.26ITT762 pKa = 3.52
MM1 pKa = 7.52EE2 pKa = 4.69NKK4 pKa = 9.92IYY6 pKa = 10.13QLPEE10 pKa = 4.32FIKK13 pKa = 10.44HH14 pKa = 4.74VPVANRR20 pKa = 11.84ADD22 pKa = 3.8YY23 pKa = 10.43IYY25 pKa = 11.13DD26 pKa = 3.56KK27 pKa = 10.87LYY29 pKa = 9.54KK30 pKa = 9.34TGSRR34 pKa = 11.84YY35 pKa = 10.2SVTDD39 pKa = 3.39HH40 pKa = 6.45SQFEE44 pKa = 4.11AHH46 pKa = 5.72FTRR49 pKa = 11.84NIMEE53 pKa = 4.9HH54 pKa = 7.33IEE56 pKa = 4.19FVVYY60 pKa = 10.16RR61 pKa = 11.84YY62 pKa = 10.23LIQNLPNKK70 pKa = 9.76EE71 pKa = 4.45DD72 pKa = 3.55YY73 pKa = 9.94EE74 pKa = 4.27QLFKK78 pKa = 11.22VLQGKK83 pKa = 7.96NKK85 pKa = 9.11CCFKK89 pKa = 10.32KK90 pKa = 10.48GHH92 pKa = 6.45YY93 pKa = 9.07IVNATRR99 pKa = 11.84MSGEE103 pKa = 4.2MNTSLGNGLTNLLLMLYY120 pKa = 10.08VLNQNGNTNVSGVVEE135 pKa = 4.31GDD137 pKa = 3.66DD138 pKa = 3.91GLFRR142 pKa = 11.84YY143 pKa = 9.67DD144 pKa = 4.92GPDD147 pKa = 3.15PTKK150 pKa = 11.51DD151 pKa = 3.03MFAKK155 pKa = 10.02MGFTVKK161 pKa = 10.9LEE163 pKa = 3.86IFDD166 pKa = 4.43NLNEE170 pKa = 4.06ASFCGLVFDD179 pKa = 4.75IDD181 pKa = 3.38ARR183 pKa = 11.84QIIINPIKK191 pKa = 10.67VLLNFGWLHH200 pKa = 5.0YY201 pKa = 9.67QYY203 pKa = 11.32KK204 pKa = 9.76DD205 pKa = 3.19AKK207 pKa = 10.16FEE209 pKa = 4.62TIMGLLRR216 pKa = 11.84MKK218 pKa = 10.39SLSLLYY224 pKa = 9.85QYY226 pKa = 9.11PSCPIVSEE234 pKa = 3.88LALYY238 pKa = 9.45GLRR241 pKa = 11.84VTSKK245 pKa = 8.4YY246 pKa = 8.93TPVLIRR252 pKa = 11.84EE253 pKa = 4.17NNYY256 pKa = 9.12KK257 pKa = 10.36RR258 pKa = 11.84KK259 pKa = 9.04FHH261 pKa = 6.63EE262 pKa = 4.38EE263 pKa = 3.68VLRR266 pKa = 11.84TPRR269 pKa = 11.84LVADD273 pKa = 4.47MINFKK278 pKa = 9.79TRR280 pKa = 11.84LVCEE284 pKa = 4.16KK285 pKa = 10.28VFKK288 pKa = 10.07ITIDD292 pKa = 3.56DD293 pKa = 3.67QLSIEE298 pKa = 4.69SYY300 pKa = 10.64LKK302 pKa = 10.39QLDD305 pKa = 4.04EE306 pKa = 5.47IKK308 pKa = 10.44PLWHH312 pKa = 6.79LSFHH316 pKa = 6.68TYY318 pKa = 8.58MNQYY322 pKa = 9.57QDD324 pKa = 4.19VYY326 pKa = 9.27LQYY329 pKa = 10.65VGEE332 pKa = 4.27YY333 pKa = 8.98AAGYY337 pKa = 7.46HH338 pKa = 4.91TVVPHH343 pKa = 6.38KK344 pKa = 9.63MVRR347 pKa = 11.84RR348 pKa = 11.84NRR350 pKa = 11.84TKK352 pKa = 9.35QQKK355 pKa = 9.22KK356 pKa = 10.23RR357 pKa = 11.84NNSNNNNNNNNNSKK371 pKa = 10.23KK372 pKa = 10.35KK373 pKa = 9.13EE374 pKa = 3.99IVVVRR379 pKa = 11.84QNNPKK384 pKa = 6.94QTKK387 pKa = 10.39SIGQQIGSALGGLIGHH403 pKa = 6.89GAHH406 pKa = 6.22MLFKK410 pKa = 10.61NITGFGDD417 pKa = 3.62YY418 pKa = 10.75NINNNTLLQGGMTPPEE434 pKa = 4.43VINTVRR440 pKa = 11.84EE441 pKa = 3.96GGFVVRR447 pKa = 11.84HH448 pKa = 5.67RR449 pKa = 11.84EE450 pKa = 4.04YY451 pKa = 10.56IGDD454 pKa = 3.9ISPSQQFSSTVYY466 pKa = 10.63SINPGNANTFPWLSQVADD484 pKa = 3.45SFEE487 pKa = 4.19QYY489 pKa = 9.7EE490 pKa = 4.08FRR492 pKa = 11.84GLVFEE497 pKa = 5.1FKK499 pKa = 9.7STSSDD504 pKa = 3.17AVLSSAASSSLGTVIMATQYY524 pKa = 11.46NSLSNPFTDD533 pKa = 3.12KK534 pKa = 10.62RR535 pKa = 11.84TMEE538 pKa = 4.08NYY540 pKa = 10.06EE541 pKa = 4.04YY542 pKa = 11.24ANSDD546 pKa = 3.6KK547 pKa = 11.03PSMSFYY553 pKa = 11.07HH554 pKa = 6.54PVEE557 pKa = 4.42CKK559 pKa = 10.36KK560 pKa = 10.76SQTSIDD566 pKa = 3.61LLYY569 pKa = 11.04VRR571 pKa = 11.84NTDD574 pKa = 3.54AVNGDD579 pKa = 3.42LRR581 pKa = 11.84LYY583 pKa = 10.94DD584 pKa = 4.34LGQFQISTTGMQNTGGVIGEE604 pKa = 4.28LWCTFEE610 pKa = 4.22IAFYY614 pKa = 10.4KK615 pKa = 10.55PKK617 pKa = 10.27LLPSGTTCLTDD628 pKa = 3.69HH629 pKa = 6.53YY630 pKa = 10.23RR631 pKa = 11.84LDD633 pKa = 5.44AITTDD638 pKa = 2.78VWFGDD643 pKa = 3.98TQTLMAGSNIGTNITTGGTRR663 pKa = 11.84IQFPATLSTGKK674 pKa = 10.32FSVVYY679 pKa = 9.73SISSITGFNTPPTISGSGLNLLPLYY704 pKa = 10.86NGGTTSTVIAAQSTLSTAIIGVTVQITDD732 pKa = 3.59PNAYY736 pKa = 10.33LDD738 pKa = 4.08FVGGAIGAAAEE749 pKa = 4.22GDD751 pKa = 3.88LFISQIALEE760 pKa = 4.26ITT762 pKa = 3.52
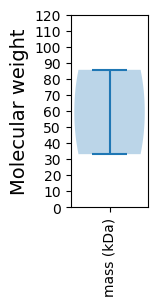
Molecular weight: 85.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFQ5|A0A1L3KFQ5_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 21 OX=1922815 PE=4 SV=1
MM1 pKa = 7.29HH2 pKa = 7.86ASPLYY7 pKa = 10.19MSMSVSKK14 pKa = 9.66TPVYY18 pKa = 10.8GSGYY22 pKa = 9.99KK23 pKa = 10.29NGIVADD29 pKa = 4.26FFFPAGYY36 pKa = 7.42TATGDD41 pKa = 3.59TTTQQCFVSYY51 pKa = 10.2EE52 pKa = 4.15LLMQLYY58 pKa = 8.26NQNYY62 pKa = 8.54LNHH65 pKa = 5.99IFEE68 pKa = 4.64RR69 pKa = 11.84SMVYY73 pKa = 10.3KK74 pKa = 10.65KK75 pKa = 10.76LINVASNCGTMNIDD89 pKa = 2.97RR90 pKa = 11.84SYY92 pKa = 11.3DD93 pKa = 3.47LKK95 pKa = 11.32YY96 pKa = 11.15DD97 pKa = 4.07LVNNTINAAIMKK109 pKa = 9.7YY110 pKa = 10.47DD111 pKa = 3.49SLVFNYY117 pKa = 10.79AEE119 pKa = 4.61TDD121 pKa = 4.2FQSHH125 pKa = 6.84PPNTEE130 pKa = 3.57SSSVGTEE137 pKa = 3.31SRR139 pKa = 11.84MPLCQNSLTQKK150 pKa = 9.48MMSSVEE156 pKa = 4.02STARR160 pKa = 11.84NCVHH164 pKa = 5.85STRR167 pKa = 11.84ALSAFLPDD175 pKa = 4.19VILKK179 pKa = 8.19GTASLYY185 pKa = 10.53QIASGSPMYY194 pKa = 10.6LVAWRR199 pKa = 11.84NASLLKK205 pKa = 10.93LPTFLMNPYY214 pKa = 9.64WILPDD219 pKa = 3.86LLITGVVQHH228 pKa = 6.77LNHH231 pKa = 6.65YY232 pKa = 9.35YY233 pKa = 7.72PTKK236 pKa = 10.36FYY238 pKa = 10.4QLKK241 pKa = 10.19SGYY244 pKa = 9.07SNQIIAVQDD253 pKa = 3.39KK254 pKa = 10.87QNFLGAIITTKK265 pKa = 10.93NLVLNSATVILALRR279 pKa = 11.84LLLNTKK285 pKa = 9.12PIWNTNGRR293 pKa = 11.84VV294 pKa = 3.12
MM1 pKa = 7.29HH2 pKa = 7.86ASPLYY7 pKa = 10.19MSMSVSKK14 pKa = 9.66TPVYY18 pKa = 10.8GSGYY22 pKa = 9.99KK23 pKa = 10.29NGIVADD29 pKa = 4.26FFFPAGYY36 pKa = 7.42TATGDD41 pKa = 3.59TTTQQCFVSYY51 pKa = 10.2EE52 pKa = 4.15LLMQLYY58 pKa = 8.26NQNYY62 pKa = 8.54LNHH65 pKa = 5.99IFEE68 pKa = 4.64RR69 pKa = 11.84SMVYY73 pKa = 10.3KK74 pKa = 10.65KK75 pKa = 10.76LINVASNCGTMNIDD89 pKa = 2.97RR90 pKa = 11.84SYY92 pKa = 11.3DD93 pKa = 3.47LKK95 pKa = 11.32YY96 pKa = 11.15DD97 pKa = 4.07LVNNTINAAIMKK109 pKa = 9.7YY110 pKa = 10.47DD111 pKa = 3.49SLVFNYY117 pKa = 10.79AEE119 pKa = 4.61TDD121 pKa = 4.2FQSHH125 pKa = 6.84PPNTEE130 pKa = 3.57SSSVGTEE137 pKa = 3.31SRR139 pKa = 11.84MPLCQNSLTQKK150 pKa = 9.48MMSSVEE156 pKa = 4.02STARR160 pKa = 11.84NCVHH164 pKa = 5.85STRR167 pKa = 11.84ALSAFLPDD175 pKa = 4.19VILKK179 pKa = 8.19GTASLYY185 pKa = 10.53QIASGSPMYY194 pKa = 10.6LVAWRR199 pKa = 11.84NASLLKK205 pKa = 10.93LPTFLMNPYY214 pKa = 9.64WILPDD219 pKa = 3.86LLITGVVQHH228 pKa = 6.77LNHH231 pKa = 6.65YY232 pKa = 9.35YY233 pKa = 7.72PTKK236 pKa = 10.36FYY238 pKa = 10.4QLKK241 pKa = 10.19SGYY244 pKa = 9.07SNQIIAVQDD253 pKa = 3.39KK254 pKa = 10.87QNFLGAIITTKK265 pKa = 10.93NLVLNSATVILALRR279 pKa = 11.84LLLNTKK285 pKa = 9.12PIWNTNGRR293 pKa = 11.84VV294 pKa = 3.12
Molecular weight: 33.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1056 |
294 |
762 |
528.0 |
59.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.114 ± 0.718 | 1.136 ± 0.119 |
4.261 ± 0.458 | 3.788 ± 0.93 |
4.64 ± 0.479 | 6.629 ± 1.175 |
2.083 ± 0.023 | 6.345 ± 0.481 |
5.398 ± 0.339 | 9.564 ± 0.884 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.936 ± 0.61 | 7.765 ± 0.212 |
3.883 ± 0.287 | 4.545 ± 0.066 |
3.409 ± 0.366 | 7.86 ± 0.886 |
7.86 ± 0.02 | 6.439 ± 0.012 |
0.758 ± 0.14 | 5.587 ± 0.466 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
