
Changjiang picorna-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
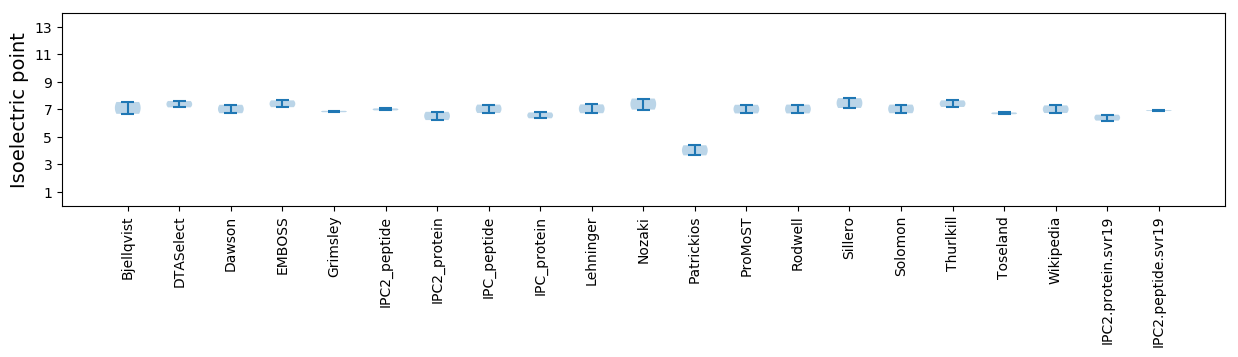
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNL4|A0A1L3KNL4_9VIRU CRPV_capsid domain-containing protein OS=Changjiang picorna-like virus 9 OX=1922798 PE=4 SV=1
MM1 pKa = 6.98VQLKK5 pKa = 10.18VPFLWHH11 pKa = 6.18YY12 pKa = 10.57EE13 pKa = 3.89FLPVQDD19 pKa = 3.63TTAGVPYY26 pKa = 10.45GLAALNVILPVPAVAGLALPTYY48 pKa = 10.25KK49 pKa = 10.1IFVHH53 pKa = 5.98MDD55 pKa = 2.99DD56 pKa = 4.63MEE58 pKa = 5.13LLGADD63 pKa = 3.56NQVATSIIPQGGKK76 pKa = 9.56FVEE79 pKa = 5.31KK80 pKa = 10.24EE81 pKa = 4.01VTDD84 pKa = 3.92NKK86 pKa = 9.98PSSVVAHH93 pKa = 6.15VSDD96 pKa = 3.83TVRR99 pKa = 11.84FLARR103 pKa = 11.84GIPAISSFTSPANWFLDD120 pKa = 3.87GVAGSLKK127 pKa = 10.51AFGYY131 pKa = 10.08SKK133 pKa = 10.39PLNLEE138 pKa = 4.26SIHH141 pKa = 6.74RR142 pKa = 11.84MQRR145 pKa = 11.84RR146 pKa = 11.84YY147 pKa = 10.45DD148 pKa = 3.48VGEE151 pKa = 4.14NNVDD155 pKa = 3.78LPVGNQLLGPLSDD168 pKa = 3.14NHH170 pKa = 7.24LPVSTFCNTDD180 pKa = 2.87VDD182 pKa = 4.23EE183 pKa = 4.9MSLKK187 pKa = 10.89YY188 pKa = 10.25ILSQYY193 pKa = 9.9NQICIGGVTPSEE205 pKa = 3.92VHH207 pKa = 5.25GTVIYY212 pKa = 7.77GTNVSPSFFWFRR224 pKa = 11.84STSLPYY230 pKa = 10.9GNWPAPLYY238 pKa = 10.11STATTNAFIPSNLFYY253 pKa = 10.31IGQMFRR259 pKa = 11.84LWRR262 pKa = 11.84GGFRR266 pKa = 11.84FRR268 pKa = 11.84VTFAKK273 pKa = 9.67TKK275 pKa = 8.5HH276 pKa = 5.9HH277 pKa = 7.1GGRR280 pKa = 11.84MMISYY285 pKa = 10.46NPTYY289 pKa = 11.14NDD291 pKa = 3.83FLTSSNTGTPVSAPEE306 pKa = 3.71ISGGLVQPFGYY317 pKa = 9.64TMICDD322 pKa = 3.75LRR324 pKa = 11.84DD325 pKa = 3.72DD326 pKa = 4.13NVFEE330 pKa = 4.52FDD332 pKa = 3.36VSYY335 pKa = 9.8WCPAPYY341 pKa = 9.35MAFDD345 pKa = 3.42SHH347 pKa = 8.42IGGLTITVMDD357 pKa = 5.25PLQAPAVVNQGVGFLVEE374 pKa = 4.81VKK376 pKa = 10.39ALPDD380 pKa = 2.91FHH382 pKa = 7.12FAVPRR387 pKa = 11.84GPRR390 pKa = 11.84YY391 pKa = 9.43PPMVGNPAIRR401 pKa = 11.84KK402 pKa = 8.67QSGKK406 pKa = 8.96MVSILKK412 pKa = 10.45EE413 pKa = 4.26SDD415 pKa = 3.45CQDD418 pKa = 4.02CIGEE422 pKa = 4.45KK423 pKa = 9.86IEE425 pKa = 4.58SVKK428 pKa = 10.14QLIMIPSRR436 pKa = 11.84TATTGINPSTFATQVIPPWFAYY458 pKa = 9.0RR459 pKa = 11.84QPDD462 pKa = 3.58TSVPFPNGFSRR473 pKa = 11.84PFSMRR478 pKa = 11.84YY479 pKa = 8.8GSSIALMYY487 pKa = 10.04TYY489 pKa = 10.2ATGSTDD495 pKa = 3.68FGIVPVTPDD504 pKa = 3.64TSPLLEE510 pKa = 5.09IGYY513 pKa = 7.88TPSDD517 pKa = 3.47EE518 pKa = 5.13GIGFAANNVQQKK530 pKa = 8.68PANSILRR537 pKa = 11.84QYY539 pKa = 11.57ARR541 pKa = 11.84GDD543 pKa = 2.93QGLFVRR549 pKa = 11.84TPAYY553 pKa = 9.85FKK555 pKa = 9.91YY556 pKa = 10.35KK557 pKa = 9.98RR558 pKa = 11.84VPVHH562 pKa = 5.32YY563 pKa = 9.27WVSGSNNFLIDD574 pKa = 3.22GTRR577 pKa = 11.84NNRR580 pKa = 11.84FSSALHH586 pKa = 4.95AQSRR590 pKa = 11.84IVQPYY595 pKa = 7.98VQITNGAVASYY606 pKa = 10.32FRR608 pKa = 11.84MYY610 pKa = 10.87LSAGDD615 pKa = 3.96DD616 pKa = 3.35ARR618 pKa = 11.84LGLFIGPPPLHH629 pKa = 6.31LTQSGYY635 pKa = 10.79SADD638 pKa = 3.7IDD640 pKa = 3.5SDD642 pKa = 4.6YY643 pKa = 11.4PVVASS648 pKa = 4.03
MM1 pKa = 6.98VQLKK5 pKa = 10.18VPFLWHH11 pKa = 6.18YY12 pKa = 10.57EE13 pKa = 3.89FLPVQDD19 pKa = 3.63TTAGVPYY26 pKa = 10.45GLAALNVILPVPAVAGLALPTYY48 pKa = 10.25KK49 pKa = 10.1IFVHH53 pKa = 5.98MDD55 pKa = 2.99DD56 pKa = 4.63MEE58 pKa = 5.13LLGADD63 pKa = 3.56NQVATSIIPQGGKK76 pKa = 9.56FVEE79 pKa = 5.31KK80 pKa = 10.24EE81 pKa = 4.01VTDD84 pKa = 3.92NKK86 pKa = 9.98PSSVVAHH93 pKa = 6.15VSDD96 pKa = 3.83TVRR99 pKa = 11.84FLARR103 pKa = 11.84GIPAISSFTSPANWFLDD120 pKa = 3.87GVAGSLKK127 pKa = 10.51AFGYY131 pKa = 10.08SKK133 pKa = 10.39PLNLEE138 pKa = 4.26SIHH141 pKa = 6.74RR142 pKa = 11.84MQRR145 pKa = 11.84RR146 pKa = 11.84YY147 pKa = 10.45DD148 pKa = 3.48VGEE151 pKa = 4.14NNVDD155 pKa = 3.78LPVGNQLLGPLSDD168 pKa = 3.14NHH170 pKa = 7.24LPVSTFCNTDD180 pKa = 2.87VDD182 pKa = 4.23EE183 pKa = 4.9MSLKK187 pKa = 10.89YY188 pKa = 10.25ILSQYY193 pKa = 9.9NQICIGGVTPSEE205 pKa = 3.92VHH207 pKa = 5.25GTVIYY212 pKa = 7.77GTNVSPSFFWFRR224 pKa = 11.84STSLPYY230 pKa = 10.9GNWPAPLYY238 pKa = 10.11STATTNAFIPSNLFYY253 pKa = 10.31IGQMFRR259 pKa = 11.84LWRR262 pKa = 11.84GGFRR266 pKa = 11.84FRR268 pKa = 11.84VTFAKK273 pKa = 9.67TKK275 pKa = 8.5HH276 pKa = 5.9HH277 pKa = 7.1GGRR280 pKa = 11.84MMISYY285 pKa = 10.46NPTYY289 pKa = 11.14NDD291 pKa = 3.83FLTSSNTGTPVSAPEE306 pKa = 3.71ISGGLVQPFGYY317 pKa = 9.64TMICDD322 pKa = 3.75LRR324 pKa = 11.84DD325 pKa = 3.72DD326 pKa = 4.13NVFEE330 pKa = 4.52FDD332 pKa = 3.36VSYY335 pKa = 9.8WCPAPYY341 pKa = 9.35MAFDD345 pKa = 3.42SHH347 pKa = 8.42IGGLTITVMDD357 pKa = 5.25PLQAPAVVNQGVGFLVEE374 pKa = 4.81VKK376 pKa = 10.39ALPDD380 pKa = 2.91FHH382 pKa = 7.12FAVPRR387 pKa = 11.84GPRR390 pKa = 11.84YY391 pKa = 9.43PPMVGNPAIRR401 pKa = 11.84KK402 pKa = 8.67QSGKK406 pKa = 8.96MVSILKK412 pKa = 10.45EE413 pKa = 4.26SDD415 pKa = 3.45CQDD418 pKa = 4.02CIGEE422 pKa = 4.45KK423 pKa = 9.86IEE425 pKa = 4.58SVKK428 pKa = 10.14QLIMIPSRR436 pKa = 11.84TATTGINPSTFATQVIPPWFAYY458 pKa = 9.0RR459 pKa = 11.84QPDD462 pKa = 3.58TSVPFPNGFSRR473 pKa = 11.84PFSMRR478 pKa = 11.84YY479 pKa = 8.8GSSIALMYY487 pKa = 10.04TYY489 pKa = 10.2ATGSTDD495 pKa = 3.68FGIVPVTPDD504 pKa = 3.64TSPLLEE510 pKa = 5.09IGYY513 pKa = 7.88TPSDD517 pKa = 3.47EE518 pKa = 5.13GIGFAANNVQQKK530 pKa = 8.68PANSILRR537 pKa = 11.84QYY539 pKa = 11.57ARR541 pKa = 11.84GDD543 pKa = 2.93QGLFVRR549 pKa = 11.84TPAYY553 pKa = 9.85FKK555 pKa = 9.91YY556 pKa = 10.35KK557 pKa = 9.98RR558 pKa = 11.84VPVHH562 pKa = 5.32YY563 pKa = 9.27WVSGSNNFLIDD574 pKa = 3.22GTRR577 pKa = 11.84NNRR580 pKa = 11.84FSSALHH586 pKa = 4.95AQSRR590 pKa = 11.84IVQPYY595 pKa = 7.98VQITNGAVASYY606 pKa = 10.32FRR608 pKa = 11.84MYY610 pKa = 10.87LSAGDD615 pKa = 3.96DD616 pKa = 3.35ARR618 pKa = 11.84LGLFIGPPPLHH629 pKa = 6.31LTQSGYY635 pKa = 10.79SADD638 pKa = 3.7IDD640 pKa = 3.5SDD642 pKa = 4.6YY643 pKa = 11.4PVVASS648 pKa = 4.03
Molecular weight: 71.28 kDa
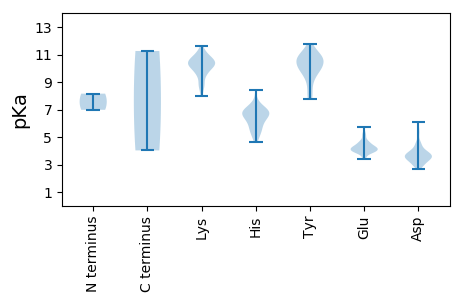
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNL4|A0A1L3KNL4_9VIRU CRPV_capsid domain-containing protein OS=Changjiang picorna-like virus 9 OX=1922798 PE=4 SV=1
MM1 pKa = 8.16DD2 pKa = 5.51KK3 pKa = 11.22LNLGTPYY10 pKa = 9.53VTVATVKK17 pKa = 9.71RR18 pKa = 11.84TISVSDD24 pKa = 3.61YY25 pKa = 11.42LDD27 pKa = 3.38VKK29 pKa = 11.03KK30 pKa = 10.52LDD32 pKa = 3.4RR33 pKa = 11.84MLALGLITISGNYY46 pKa = 8.26GKK48 pKa = 10.52SYY50 pKa = 11.03LNGGRR55 pKa = 11.84PQLSEE60 pKa = 4.22HH61 pKa = 7.06LDD63 pKa = 3.52RR64 pKa = 11.84DD65 pKa = 3.49CRR67 pKa = 11.84YY68 pKa = 7.88YY69 pKa = 10.91TEE71 pKa = 3.84VRR73 pKa = 11.84TYY75 pKa = 10.54FATLLILWDD84 pKa = 4.04YY85 pKa = 11.41VNVIPAAKK93 pKa = 9.82RR94 pKa = 11.84ANLCEE99 pKa = 4.06ARR101 pKa = 11.84EE102 pKa = 4.1QSTIDD107 pKa = 3.08HH108 pKa = 6.82KK109 pKa = 10.83FRR111 pKa = 11.84RR112 pKa = 11.84LQRR115 pKa = 11.84DD116 pKa = 2.81HH117 pKa = 7.43DD118 pKa = 3.74RR119 pKa = 11.84RR120 pKa = 11.84NFSKK124 pKa = 10.67SRR126 pKa = 11.84EE127 pKa = 4.11TPAMRR132 pKa = 11.84RR133 pKa = 11.84SRR135 pKa = 11.84DD136 pKa = 3.15RR137 pKa = 11.84QLQDD141 pKa = 2.78RR142 pKa = 11.84RR143 pKa = 11.84SEE145 pKa = 4.05KK146 pKa = 10.54DD147 pKa = 2.88SRR149 pKa = 11.84TAYY152 pKa = 9.97FAGVTEE158 pKa = 4.15QAMGRR163 pKa = 11.84LLMGSDD169 pKa = 2.83NVKK172 pKa = 10.21RR173 pKa = 11.84VEE175 pKa = 3.98KK176 pKa = 10.49LVEE179 pKa = 4.0NLEE182 pKa = 4.38TLTSKK187 pKa = 10.97ASPAIDD193 pKa = 3.57AGVDD197 pKa = 4.51LISLIHH203 pKa = 6.46KK204 pKa = 9.72GIEE207 pKa = 4.27SMVATLKK214 pKa = 9.07QTLGFMLWAVPVSLLLYY231 pKa = 9.14YY232 pKa = 9.97IYY234 pKa = 10.65RR235 pKa = 11.84RR236 pKa = 11.84FGVTSAVAGLAIIGVARR253 pKa = 11.84AILPKK258 pKa = 10.61DD259 pKa = 3.36LWTKK263 pKa = 10.22LHH265 pKa = 6.11TFFTPAEE272 pKa = 4.18EE273 pKa = 3.93QSNMNFMSHH282 pKa = 6.64ILCSTASFVFLRR294 pKa = 11.84GKK296 pKa = 9.1TSADD300 pKa = 3.42VVDD303 pKa = 4.64DD304 pKa = 4.64LLRR307 pKa = 11.84KK308 pKa = 9.19LANLPRR314 pKa = 11.84AVTGLEE320 pKa = 3.97GFLEE324 pKa = 4.49FMKK327 pKa = 10.48KK328 pKa = 10.22GVMQVVEE335 pKa = 5.39FFAKK339 pKa = 10.27RR340 pKa = 11.84FHH342 pKa = 6.87PNLAKK347 pKa = 10.61RR348 pKa = 11.84MEE350 pKa = 4.26KK351 pKa = 10.47LRR353 pKa = 11.84DD354 pKa = 3.67PVDD357 pKa = 3.04VWIRR361 pKa = 11.84KK362 pKa = 8.85IDD364 pKa = 3.8EE365 pKa = 4.73LEE367 pKa = 4.14LASVTNQSDD376 pKa = 3.95PDD378 pKa = 3.69QKK380 pKa = 11.65ALDD383 pKa = 4.27EE384 pKa = 5.34LVDD387 pKa = 3.96CCVQGAAYY395 pKa = 10.12RR396 pKa = 11.84DD397 pKa = 4.09AFRR400 pKa = 11.84HH401 pKa = 4.66TPMEE405 pKa = 4.05KK406 pKa = 10.0FICEE410 pKa = 3.96AFAKK414 pKa = 10.14AKK416 pKa = 10.34RR417 pKa = 11.84LLSPHH422 pKa = 6.43TGALSARR429 pKa = 11.84NNFRR433 pKa = 11.84VEE435 pKa = 3.99PVSVFLYY442 pKa = 10.71GGAGVGKK449 pKa = 8.76TSILTRR455 pKa = 11.84FATSALVMSGLCEE468 pKa = 4.43AKK470 pKa = 10.16DD471 pKa = 3.44AKK473 pKa = 11.1ANLWQKK479 pKa = 8.7GTSKK483 pKa = 10.6YY484 pKa = 7.78WNSYY488 pKa = 9.64AGQLALGLDD497 pKa = 4.0DD498 pKa = 6.11FGQEE502 pKa = 4.28KK503 pKa = 9.94PYY505 pKa = 10.5QGQEE509 pKa = 3.47EE510 pKa = 4.74SEE512 pKa = 4.43YY513 pKa = 8.5MTLIRR518 pKa = 11.84AKK520 pKa = 10.08GCWSFPLAMADD531 pKa = 3.64VEE533 pKa = 5.14SKK535 pKa = 11.02GKK537 pKa = 8.23TFFRR541 pKa = 11.84SKK543 pKa = 11.2LMVATTNVDD552 pKa = 2.95KK553 pKa = 11.12MKK555 pKa = 10.69DD556 pKa = 3.35QVGTILYY563 pKa = 9.75SDD565 pKa = 3.79SAVEE569 pKa = 4.25RR570 pKa = 11.84RR571 pKa = 11.84IDD573 pKa = 3.4YY574 pKa = 9.28PLRR577 pKa = 11.84LEE579 pKa = 3.93VHH581 pKa = 6.7PDD583 pKa = 3.25YY584 pKa = 11.02LKK586 pKa = 10.85EE587 pKa = 4.26GGVPGEE593 pKa = 4.04LDD595 pKa = 3.39YY596 pKa = 11.76EE597 pKa = 4.26KK598 pKa = 10.7LQKK601 pKa = 10.23EE602 pKa = 4.53VARR605 pKa = 11.84RR606 pKa = 11.84AQLYY610 pKa = 10.23RR611 pKa = 11.84DD612 pKa = 3.48GKK614 pKa = 8.81LTLRR618 pKa = 11.84SPCDD622 pKa = 3.43VYY624 pKa = 10.73PWYY627 pKa = 10.01VWKK630 pKa = 10.63VYY632 pKa = 10.52KK633 pKa = 10.3HH634 pKa = 6.82DD635 pKa = 3.96FFSVQTGEE643 pKa = 3.92PVEE646 pKa = 4.24YY647 pKa = 10.19DD648 pKa = 3.21LHH650 pKa = 6.93DD651 pKa = 4.58LLVEE655 pKa = 4.23ISKK658 pKa = 10.77KK659 pKa = 10.58LRR661 pKa = 11.84DD662 pKa = 3.5RR663 pKa = 11.84SAMHH667 pKa = 6.6KK668 pKa = 10.36NAEE671 pKa = 3.98ADD673 pKa = 3.67LDD675 pKa = 4.15DD676 pKa = 5.26FLAGLTPIKK685 pKa = 9.49EE686 pKa = 3.92QSGRR690 pKa = 11.84EE691 pKa = 3.78IEE693 pKa = 4.59EE694 pKa = 3.54ISTFLDD700 pKa = 3.69GDD702 pKa = 3.92FLDD705 pKa = 4.57LDD707 pKa = 5.01EE708 pKa = 5.74ISEE711 pKa = 4.18MTLEE715 pKa = 4.0QFAEE719 pKa = 3.97QRR721 pKa = 11.84EE722 pKa = 4.32KK723 pKa = 11.43VEE725 pKa = 3.93TSLRR729 pKa = 11.84MHH731 pKa = 6.67TMSKK735 pKa = 8.01TLKK738 pKa = 10.47FQDD741 pKa = 2.81RR742 pKa = 11.84WEE744 pKa = 4.23LFKK747 pKa = 11.17LWINTQITSVAAVIHH762 pKa = 5.62RR763 pKa = 11.84NKK765 pKa = 9.84WIILGVVAVAAAVGLLIAAVRR786 pKa = 11.84GMWSFVKK793 pKa = 10.67SFFSKK798 pKa = 10.72KK799 pKa = 9.64KK800 pKa = 10.82DD801 pKa = 3.18NDD803 pKa = 3.24KK804 pKa = 10.89GQRR807 pKa = 11.84RR808 pKa = 11.84QSNRR812 pKa = 11.84LVTKK816 pKa = 9.33PRR818 pKa = 11.84PVVQLGSDD826 pKa = 3.53NAVLRR831 pKa = 11.84TFYY834 pKa = 11.24SDD836 pKa = 3.26TYY838 pKa = 11.26KK839 pKa = 10.36MFVEE843 pKa = 4.37YY844 pKa = 10.83VVGGEE849 pKa = 4.01ALCEE853 pKa = 3.8IFGQVQFLNGGISVCPYY870 pKa = 10.61HH871 pKa = 7.1FITSLEE877 pKa = 4.06SLEE880 pKa = 4.14KK881 pKa = 10.81SKK883 pKa = 11.45DD884 pKa = 3.14VGTEE888 pKa = 3.37VRR890 pKa = 11.84VNMRR894 pKa = 11.84NAINGDD900 pKa = 3.86LEE902 pKa = 4.21YY903 pKa = 10.95GAAKK907 pKa = 10.2AVTAAQILKK916 pKa = 10.09LPRR919 pKa = 11.84EE920 pKa = 4.2RR921 pKa = 11.84VPGEE925 pKa = 3.85DD926 pKa = 3.11VVFMNWEE933 pKa = 4.0PLNVRR938 pKa = 11.84AHH940 pKa = 5.35TAKK943 pKa = 9.17EE944 pKa = 3.49HH945 pKa = 6.38HH946 pKa = 7.04IIEE949 pKa = 4.43EE950 pKa = 4.13EE951 pKa = 4.11HH952 pKa = 6.71FKK954 pKa = 10.74LLSMASRR961 pKa = 11.84PARR964 pKa = 11.84LSICDD969 pKa = 3.15IDD971 pKa = 3.92INGVLQSGTMPRR983 pKa = 11.84FFQCEE988 pKa = 3.72KK989 pKa = 10.83LIAHH993 pKa = 6.86KK994 pKa = 10.32PMRR997 pKa = 11.84LTNSSEE1003 pKa = 3.89NARR1006 pKa = 11.84VWEE1009 pKa = 4.22YY1010 pKa = 11.43CLNTQEE1016 pKa = 4.71GDD1018 pKa = 3.42CGAPLTLTKK1027 pKa = 10.24QDD1029 pKa = 3.63GVGKK1033 pKa = 10.23YY1034 pKa = 9.71YY1035 pKa = 10.81LGFHH1039 pKa = 6.94IAGDD1043 pKa = 3.67GHH1045 pKa = 6.42NKK1047 pKa = 9.43GFSAVVTKK1055 pKa = 10.58EE1056 pKa = 3.41MYY1058 pKa = 10.11DD1059 pKa = 2.95RR1060 pKa = 11.84HH1061 pKa = 4.82TAKK1064 pKa = 10.57LGVKK1068 pKa = 9.46GRR1070 pKa = 11.84PVVKK1074 pKa = 9.52DD1075 pKa = 3.36TLMPACEE1082 pKa = 3.94QAGRR1086 pKa = 11.84PILEE1090 pKa = 4.65FEE1092 pKa = 4.61DD1093 pKa = 3.56TDD1095 pKa = 5.33KK1096 pKa = 11.33IPEE1099 pKa = 4.46AYY1101 pKa = 10.13NSGSFTVIGKK1111 pKa = 8.03VEE1113 pKa = 3.94KK1114 pKa = 10.49AVNVSSKK1121 pKa = 11.12SKK1123 pKa = 10.11IFPTVLHH1130 pKa = 6.37GVLGPAVDD1138 pKa = 3.82VPVALGPVRR1147 pKa = 11.84DD1148 pKa = 4.23GAHH1151 pKa = 6.72TIYY1154 pKa = 10.64PMEE1157 pKa = 4.63NAIQANKK1164 pKa = 8.31TPVMVVDD1171 pKa = 3.93TDD1173 pKa = 4.14LPIWQQAMHH1182 pKa = 5.77MAGRR1186 pKa = 11.84KK1187 pKa = 8.52FMEE1190 pKa = 4.06VTGPSRR1196 pKa = 11.84LNARR1200 pKa = 11.84VLTFEE1205 pKa = 4.61EE1206 pKa = 4.42ACIGNPILKK1215 pKa = 10.23LKK1217 pKa = 10.51AINRR1221 pKa = 11.84NSSAGYY1227 pKa = 8.5PYY1229 pKa = 10.58CLEE1232 pKa = 3.91YY1233 pKa = 11.1SSGKK1237 pKa = 9.63KK1238 pKa = 9.91HH1239 pKa = 5.58FFGRR1243 pKa = 11.84GDD1245 pKa = 3.67EE1246 pKa = 4.21YY1247 pKa = 11.57DD1248 pKa = 3.5LTGPAAQLLRR1258 pKa = 11.84ADD1260 pKa = 3.61VQKK1263 pKa = 10.73IIEE1266 pKa = 4.19KK1267 pKa = 9.78AKK1269 pKa = 9.98KK1270 pKa = 9.75GEE1272 pKa = 4.09RR1273 pKa = 11.84SPVIYY1278 pKa = 10.69NDD1280 pKa = 3.16FLKK1283 pKa = 10.68DD1284 pKa = 3.44EE1285 pKa = 4.38RR1286 pKa = 11.84RR1287 pKa = 11.84SPEE1290 pKa = 3.47KK1291 pKa = 10.45VANHH1295 pKa = 5.4MARR1298 pKa = 11.84LISSSPLDD1306 pKa = 3.33YY1307 pKa = 10.71TIAVRR1312 pKa = 11.84MYY1314 pKa = 9.84FGAILSAMFSNSIDD1328 pKa = 3.42SGLAPGACVYY1338 pKa = 10.76QDD1340 pKa = 2.67WGTLAKK1346 pKa = 10.17HH1347 pKa = 5.73LQSKK1351 pKa = 10.2GDD1353 pKa = 3.61AVFDD1357 pKa = 3.97GDD1359 pKa = 4.48FKK1361 pKa = 11.35RR1362 pKa = 11.84FDD1364 pKa = 3.5STQQPDD1370 pKa = 4.43LLMLILDD1377 pKa = 5.78FINDD1381 pKa = 3.63WYY1383 pKa = 11.01SVNGVPVGPHH1393 pKa = 5.67EE1394 pKa = 4.33CLEE1397 pKa = 4.41GGICEE1402 pKa = 4.59DD1403 pKa = 3.35NLVRR1407 pKa = 11.84YY1408 pKa = 9.55VLFQDD1413 pKa = 5.52LIHH1416 pKa = 6.32SRR1418 pKa = 11.84HH1419 pKa = 6.04LGGLSSKK1426 pKa = 10.73HH1427 pKa = 6.58DD1428 pKa = 4.39IVYY1431 pKa = 9.74QWNKK1435 pKa = 9.6CLPSGHH1441 pKa = 7.44PLTTLVNSLFSLLMLSYY1458 pKa = 10.73SYY1460 pKa = 10.91IVLTGDD1466 pKa = 4.11RR1467 pKa = 11.84LGFWDD1472 pKa = 3.75HH1473 pKa = 6.81CFANSFGDD1481 pKa = 3.89DD1482 pKa = 3.71NIVNVDD1488 pKa = 4.35DD1489 pKa = 3.78EE1490 pKa = 4.78TKK1492 pKa = 10.95DD1493 pKa = 3.18VFNQVTVAKK1502 pKa = 9.98VLWEE1506 pKa = 4.21DD1507 pKa = 3.01LGMVYY1512 pKa = 9.13TAGAKK1517 pKa = 10.04DD1518 pKa = 3.75GSEE1521 pKa = 3.82IPYY1524 pKa = 8.6KK1525 pKa = 10.22TLEE1528 pKa = 3.87EE1529 pKa = 4.25CTFLKK1534 pKa = 10.52RR1535 pKa = 11.84SFHH1538 pKa = 7.73FDD1540 pKa = 3.76GKK1542 pKa = 9.5WLAPLEE1548 pKa = 4.72LNSFMQTFYY1557 pKa = 10.09WAKK1560 pKa = 10.16NKK1562 pKa = 10.31LFVDD1566 pKa = 3.17QTIRR1570 pKa = 11.84SDD1572 pKa = 3.63LNNALEE1578 pKa = 4.21EE1579 pKa = 4.48LSLHH1583 pKa = 5.2TQEE1586 pKa = 5.08VWDD1589 pKa = 3.87EE1590 pKa = 3.87HH1591 pKa = 7.19APRR1594 pKa = 11.84LMQAARR1600 pKa = 11.84KK1601 pKa = 8.85FDD1603 pKa = 3.68PCFTPNAPLTLAAHH1617 pKa = 6.75RR1618 pKa = 11.84DD1619 pKa = 4.22VVCSRR1624 pKa = 11.84TDD1626 pKa = 2.77WYY1628 pKa = 11.26
MM1 pKa = 8.16DD2 pKa = 5.51KK3 pKa = 11.22LNLGTPYY10 pKa = 9.53VTVATVKK17 pKa = 9.71RR18 pKa = 11.84TISVSDD24 pKa = 3.61YY25 pKa = 11.42LDD27 pKa = 3.38VKK29 pKa = 11.03KK30 pKa = 10.52LDD32 pKa = 3.4RR33 pKa = 11.84MLALGLITISGNYY46 pKa = 8.26GKK48 pKa = 10.52SYY50 pKa = 11.03LNGGRR55 pKa = 11.84PQLSEE60 pKa = 4.22HH61 pKa = 7.06LDD63 pKa = 3.52RR64 pKa = 11.84DD65 pKa = 3.49CRR67 pKa = 11.84YY68 pKa = 7.88YY69 pKa = 10.91TEE71 pKa = 3.84VRR73 pKa = 11.84TYY75 pKa = 10.54FATLLILWDD84 pKa = 4.04YY85 pKa = 11.41VNVIPAAKK93 pKa = 9.82RR94 pKa = 11.84ANLCEE99 pKa = 4.06ARR101 pKa = 11.84EE102 pKa = 4.1QSTIDD107 pKa = 3.08HH108 pKa = 6.82KK109 pKa = 10.83FRR111 pKa = 11.84RR112 pKa = 11.84LQRR115 pKa = 11.84DD116 pKa = 2.81HH117 pKa = 7.43DD118 pKa = 3.74RR119 pKa = 11.84RR120 pKa = 11.84NFSKK124 pKa = 10.67SRR126 pKa = 11.84EE127 pKa = 4.11TPAMRR132 pKa = 11.84RR133 pKa = 11.84SRR135 pKa = 11.84DD136 pKa = 3.15RR137 pKa = 11.84QLQDD141 pKa = 2.78RR142 pKa = 11.84RR143 pKa = 11.84SEE145 pKa = 4.05KK146 pKa = 10.54DD147 pKa = 2.88SRR149 pKa = 11.84TAYY152 pKa = 9.97FAGVTEE158 pKa = 4.15QAMGRR163 pKa = 11.84LLMGSDD169 pKa = 2.83NVKK172 pKa = 10.21RR173 pKa = 11.84VEE175 pKa = 3.98KK176 pKa = 10.49LVEE179 pKa = 4.0NLEE182 pKa = 4.38TLTSKK187 pKa = 10.97ASPAIDD193 pKa = 3.57AGVDD197 pKa = 4.51LISLIHH203 pKa = 6.46KK204 pKa = 9.72GIEE207 pKa = 4.27SMVATLKK214 pKa = 9.07QTLGFMLWAVPVSLLLYY231 pKa = 9.14YY232 pKa = 9.97IYY234 pKa = 10.65RR235 pKa = 11.84RR236 pKa = 11.84FGVTSAVAGLAIIGVARR253 pKa = 11.84AILPKK258 pKa = 10.61DD259 pKa = 3.36LWTKK263 pKa = 10.22LHH265 pKa = 6.11TFFTPAEE272 pKa = 4.18EE273 pKa = 3.93QSNMNFMSHH282 pKa = 6.64ILCSTASFVFLRR294 pKa = 11.84GKK296 pKa = 9.1TSADD300 pKa = 3.42VVDD303 pKa = 4.64DD304 pKa = 4.64LLRR307 pKa = 11.84KK308 pKa = 9.19LANLPRR314 pKa = 11.84AVTGLEE320 pKa = 3.97GFLEE324 pKa = 4.49FMKK327 pKa = 10.48KK328 pKa = 10.22GVMQVVEE335 pKa = 5.39FFAKK339 pKa = 10.27RR340 pKa = 11.84FHH342 pKa = 6.87PNLAKK347 pKa = 10.61RR348 pKa = 11.84MEE350 pKa = 4.26KK351 pKa = 10.47LRR353 pKa = 11.84DD354 pKa = 3.67PVDD357 pKa = 3.04VWIRR361 pKa = 11.84KK362 pKa = 8.85IDD364 pKa = 3.8EE365 pKa = 4.73LEE367 pKa = 4.14LASVTNQSDD376 pKa = 3.95PDD378 pKa = 3.69QKK380 pKa = 11.65ALDD383 pKa = 4.27EE384 pKa = 5.34LVDD387 pKa = 3.96CCVQGAAYY395 pKa = 10.12RR396 pKa = 11.84DD397 pKa = 4.09AFRR400 pKa = 11.84HH401 pKa = 4.66TPMEE405 pKa = 4.05KK406 pKa = 10.0FICEE410 pKa = 3.96AFAKK414 pKa = 10.14AKK416 pKa = 10.34RR417 pKa = 11.84LLSPHH422 pKa = 6.43TGALSARR429 pKa = 11.84NNFRR433 pKa = 11.84VEE435 pKa = 3.99PVSVFLYY442 pKa = 10.71GGAGVGKK449 pKa = 8.76TSILTRR455 pKa = 11.84FATSALVMSGLCEE468 pKa = 4.43AKK470 pKa = 10.16DD471 pKa = 3.44AKK473 pKa = 11.1ANLWQKK479 pKa = 8.7GTSKK483 pKa = 10.6YY484 pKa = 7.78WNSYY488 pKa = 9.64AGQLALGLDD497 pKa = 4.0DD498 pKa = 6.11FGQEE502 pKa = 4.28KK503 pKa = 9.94PYY505 pKa = 10.5QGQEE509 pKa = 3.47EE510 pKa = 4.74SEE512 pKa = 4.43YY513 pKa = 8.5MTLIRR518 pKa = 11.84AKK520 pKa = 10.08GCWSFPLAMADD531 pKa = 3.64VEE533 pKa = 5.14SKK535 pKa = 11.02GKK537 pKa = 8.23TFFRR541 pKa = 11.84SKK543 pKa = 11.2LMVATTNVDD552 pKa = 2.95KK553 pKa = 11.12MKK555 pKa = 10.69DD556 pKa = 3.35QVGTILYY563 pKa = 9.75SDD565 pKa = 3.79SAVEE569 pKa = 4.25RR570 pKa = 11.84RR571 pKa = 11.84IDD573 pKa = 3.4YY574 pKa = 9.28PLRR577 pKa = 11.84LEE579 pKa = 3.93VHH581 pKa = 6.7PDD583 pKa = 3.25YY584 pKa = 11.02LKK586 pKa = 10.85EE587 pKa = 4.26GGVPGEE593 pKa = 4.04LDD595 pKa = 3.39YY596 pKa = 11.76EE597 pKa = 4.26KK598 pKa = 10.7LQKK601 pKa = 10.23EE602 pKa = 4.53VARR605 pKa = 11.84RR606 pKa = 11.84AQLYY610 pKa = 10.23RR611 pKa = 11.84DD612 pKa = 3.48GKK614 pKa = 8.81LTLRR618 pKa = 11.84SPCDD622 pKa = 3.43VYY624 pKa = 10.73PWYY627 pKa = 10.01VWKK630 pKa = 10.63VYY632 pKa = 10.52KK633 pKa = 10.3HH634 pKa = 6.82DD635 pKa = 3.96FFSVQTGEE643 pKa = 3.92PVEE646 pKa = 4.24YY647 pKa = 10.19DD648 pKa = 3.21LHH650 pKa = 6.93DD651 pKa = 4.58LLVEE655 pKa = 4.23ISKK658 pKa = 10.77KK659 pKa = 10.58LRR661 pKa = 11.84DD662 pKa = 3.5RR663 pKa = 11.84SAMHH667 pKa = 6.6KK668 pKa = 10.36NAEE671 pKa = 3.98ADD673 pKa = 3.67LDD675 pKa = 4.15DD676 pKa = 5.26FLAGLTPIKK685 pKa = 9.49EE686 pKa = 3.92QSGRR690 pKa = 11.84EE691 pKa = 3.78IEE693 pKa = 4.59EE694 pKa = 3.54ISTFLDD700 pKa = 3.69GDD702 pKa = 3.92FLDD705 pKa = 4.57LDD707 pKa = 5.01EE708 pKa = 5.74ISEE711 pKa = 4.18MTLEE715 pKa = 4.0QFAEE719 pKa = 3.97QRR721 pKa = 11.84EE722 pKa = 4.32KK723 pKa = 11.43VEE725 pKa = 3.93TSLRR729 pKa = 11.84MHH731 pKa = 6.67TMSKK735 pKa = 8.01TLKK738 pKa = 10.47FQDD741 pKa = 2.81RR742 pKa = 11.84WEE744 pKa = 4.23LFKK747 pKa = 11.17LWINTQITSVAAVIHH762 pKa = 5.62RR763 pKa = 11.84NKK765 pKa = 9.84WIILGVVAVAAAVGLLIAAVRR786 pKa = 11.84GMWSFVKK793 pKa = 10.67SFFSKK798 pKa = 10.72KK799 pKa = 9.64KK800 pKa = 10.82DD801 pKa = 3.18NDD803 pKa = 3.24KK804 pKa = 10.89GQRR807 pKa = 11.84RR808 pKa = 11.84QSNRR812 pKa = 11.84LVTKK816 pKa = 9.33PRR818 pKa = 11.84PVVQLGSDD826 pKa = 3.53NAVLRR831 pKa = 11.84TFYY834 pKa = 11.24SDD836 pKa = 3.26TYY838 pKa = 11.26KK839 pKa = 10.36MFVEE843 pKa = 4.37YY844 pKa = 10.83VVGGEE849 pKa = 4.01ALCEE853 pKa = 3.8IFGQVQFLNGGISVCPYY870 pKa = 10.61HH871 pKa = 7.1FITSLEE877 pKa = 4.06SLEE880 pKa = 4.14KK881 pKa = 10.81SKK883 pKa = 11.45DD884 pKa = 3.14VGTEE888 pKa = 3.37VRR890 pKa = 11.84VNMRR894 pKa = 11.84NAINGDD900 pKa = 3.86LEE902 pKa = 4.21YY903 pKa = 10.95GAAKK907 pKa = 10.2AVTAAQILKK916 pKa = 10.09LPRR919 pKa = 11.84EE920 pKa = 4.2RR921 pKa = 11.84VPGEE925 pKa = 3.85DD926 pKa = 3.11VVFMNWEE933 pKa = 4.0PLNVRR938 pKa = 11.84AHH940 pKa = 5.35TAKK943 pKa = 9.17EE944 pKa = 3.49HH945 pKa = 6.38HH946 pKa = 7.04IIEE949 pKa = 4.43EE950 pKa = 4.13EE951 pKa = 4.11HH952 pKa = 6.71FKK954 pKa = 10.74LLSMASRR961 pKa = 11.84PARR964 pKa = 11.84LSICDD969 pKa = 3.15IDD971 pKa = 3.92INGVLQSGTMPRR983 pKa = 11.84FFQCEE988 pKa = 3.72KK989 pKa = 10.83LIAHH993 pKa = 6.86KK994 pKa = 10.32PMRR997 pKa = 11.84LTNSSEE1003 pKa = 3.89NARR1006 pKa = 11.84VWEE1009 pKa = 4.22YY1010 pKa = 11.43CLNTQEE1016 pKa = 4.71GDD1018 pKa = 3.42CGAPLTLTKK1027 pKa = 10.24QDD1029 pKa = 3.63GVGKK1033 pKa = 10.23YY1034 pKa = 9.71YY1035 pKa = 10.81LGFHH1039 pKa = 6.94IAGDD1043 pKa = 3.67GHH1045 pKa = 6.42NKK1047 pKa = 9.43GFSAVVTKK1055 pKa = 10.58EE1056 pKa = 3.41MYY1058 pKa = 10.11DD1059 pKa = 2.95RR1060 pKa = 11.84HH1061 pKa = 4.82TAKK1064 pKa = 10.57LGVKK1068 pKa = 9.46GRR1070 pKa = 11.84PVVKK1074 pKa = 9.52DD1075 pKa = 3.36TLMPACEE1082 pKa = 3.94QAGRR1086 pKa = 11.84PILEE1090 pKa = 4.65FEE1092 pKa = 4.61DD1093 pKa = 3.56TDD1095 pKa = 5.33KK1096 pKa = 11.33IPEE1099 pKa = 4.46AYY1101 pKa = 10.13NSGSFTVIGKK1111 pKa = 8.03VEE1113 pKa = 3.94KK1114 pKa = 10.49AVNVSSKK1121 pKa = 11.12SKK1123 pKa = 10.11IFPTVLHH1130 pKa = 6.37GVLGPAVDD1138 pKa = 3.82VPVALGPVRR1147 pKa = 11.84DD1148 pKa = 4.23GAHH1151 pKa = 6.72TIYY1154 pKa = 10.64PMEE1157 pKa = 4.63NAIQANKK1164 pKa = 8.31TPVMVVDD1171 pKa = 3.93TDD1173 pKa = 4.14LPIWQQAMHH1182 pKa = 5.77MAGRR1186 pKa = 11.84KK1187 pKa = 8.52FMEE1190 pKa = 4.06VTGPSRR1196 pKa = 11.84LNARR1200 pKa = 11.84VLTFEE1205 pKa = 4.61EE1206 pKa = 4.42ACIGNPILKK1215 pKa = 10.23LKK1217 pKa = 10.51AINRR1221 pKa = 11.84NSSAGYY1227 pKa = 8.5PYY1229 pKa = 10.58CLEE1232 pKa = 3.91YY1233 pKa = 11.1SSGKK1237 pKa = 9.63KK1238 pKa = 9.91HH1239 pKa = 5.58FFGRR1243 pKa = 11.84GDD1245 pKa = 3.67EE1246 pKa = 4.21YY1247 pKa = 11.57DD1248 pKa = 3.5LTGPAAQLLRR1258 pKa = 11.84ADD1260 pKa = 3.61VQKK1263 pKa = 10.73IIEE1266 pKa = 4.19KK1267 pKa = 9.78AKK1269 pKa = 9.98KK1270 pKa = 9.75GEE1272 pKa = 4.09RR1273 pKa = 11.84SPVIYY1278 pKa = 10.69NDD1280 pKa = 3.16FLKK1283 pKa = 10.68DD1284 pKa = 3.44EE1285 pKa = 4.38RR1286 pKa = 11.84RR1287 pKa = 11.84SPEE1290 pKa = 3.47KK1291 pKa = 10.45VANHH1295 pKa = 5.4MARR1298 pKa = 11.84LISSSPLDD1306 pKa = 3.33YY1307 pKa = 10.71TIAVRR1312 pKa = 11.84MYY1314 pKa = 9.84FGAILSAMFSNSIDD1328 pKa = 3.42SGLAPGACVYY1338 pKa = 10.76QDD1340 pKa = 2.67WGTLAKK1346 pKa = 10.17HH1347 pKa = 5.73LQSKK1351 pKa = 10.2GDD1353 pKa = 3.61AVFDD1357 pKa = 3.97GDD1359 pKa = 4.48FKK1361 pKa = 11.35RR1362 pKa = 11.84FDD1364 pKa = 3.5STQQPDD1370 pKa = 4.43LLMLILDD1377 pKa = 5.78FINDD1381 pKa = 3.63WYY1383 pKa = 11.01SVNGVPVGPHH1393 pKa = 5.67EE1394 pKa = 4.33CLEE1397 pKa = 4.41GGICEE1402 pKa = 4.59DD1403 pKa = 3.35NLVRR1407 pKa = 11.84YY1408 pKa = 9.55VLFQDD1413 pKa = 5.52LIHH1416 pKa = 6.32SRR1418 pKa = 11.84HH1419 pKa = 6.04LGGLSSKK1426 pKa = 10.73HH1427 pKa = 6.58DD1428 pKa = 4.39IVYY1431 pKa = 9.74QWNKK1435 pKa = 9.6CLPSGHH1441 pKa = 7.44PLTTLVNSLFSLLMLSYY1458 pKa = 10.73SYY1460 pKa = 10.91IVLTGDD1466 pKa = 4.11RR1467 pKa = 11.84LGFWDD1472 pKa = 3.75HH1473 pKa = 6.81CFANSFGDD1481 pKa = 3.89DD1482 pKa = 3.71NIVNVDD1488 pKa = 4.35DD1489 pKa = 3.78EE1490 pKa = 4.78TKK1492 pKa = 10.95DD1493 pKa = 3.18VFNQVTVAKK1502 pKa = 9.98VLWEE1506 pKa = 4.21DD1507 pKa = 3.01LGMVYY1512 pKa = 9.13TAGAKK1517 pKa = 10.04DD1518 pKa = 3.75GSEE1521 pKa = 3.82IPYY1524 pKa = 8.6KK1525 pKa = 10.22TLEE1528 pKa = 3.87EE1529 pKa = 4.25CTFLKK1534 pKa = 10.52RR1535 pKa = 11.84SFHH1538 pKa = 7.73FDD1540 pKa = 3.76GKK1542 pKa = 9.5WLAPLEE1548 pKa = 4.72LNSFMQTFYY1557 pKa = 10.09WAKK1560 pKa = 10.16NKK1562 pKa = 10.31LFVDD1566 pKa = 3.17QTIRR1570 pKa = 11.84SDD1572 pKa = 3.63LNNALEE1578 pKa = 4.21EE1579 pKa = 4.48LSLHH1583 pKa = 5.2TQEE1586 pKa = 5.08VWDD1589 pKa = 3.87EE1590 pKa = 3.87HH1591 pKa = 7.19APRR1594 pKa = 11.84LMQAARR1600 pKa = 11.84KK1601 pKa = 8.85FDD1603 pKa = 3.68PCFTPNAPLTLAAHH1617 pKa = 6.75RR1618 pKa = 11.84DD1619 pKa = 4.22VVCSRR1624 pKa = 11.84TDD1626 pKa = 2.77WYY1628 pKa = 11.26
Molecular weight: 183.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2276 |
648 |
1628 |
1138.0 |
127.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.425 ± 0.329 | 1.406 ± 0.248 |
6.107 ± 0.525 | 4.965 ± 1.292 |
5.053 ± 0.5 | 6.854 ± 0.606 |
2.329 ± 0.167 | 4.701 ± 0.442 |
5.58 ± 1.37 | 9.315 ± 1.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.68 ± 0.029 | 3.998 ± 0.407 |
5.185 ± 1.63 | 3.427 ± 0.223 |
5.448 ± 0.583 | 6.898 ± 0.743 |
5.58 ± 0.387 | 7.821 ± 0.265 |
1.45 ± 0.111 | 3.779 ± 0.52 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
