
Avian paramyxovirus 14
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Metaavulavirus; Avian metaavulavirus 14
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
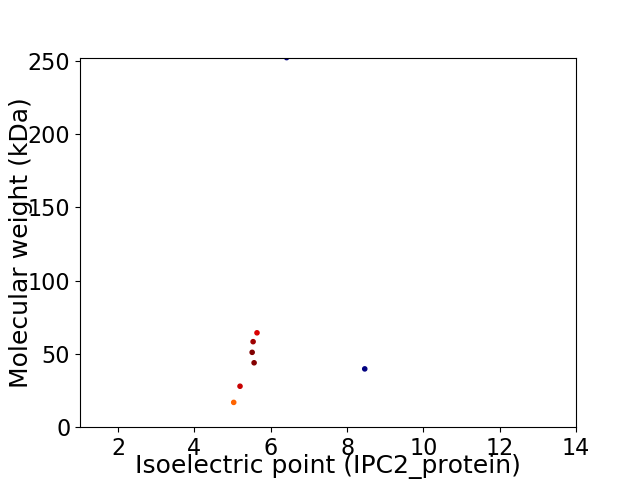
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
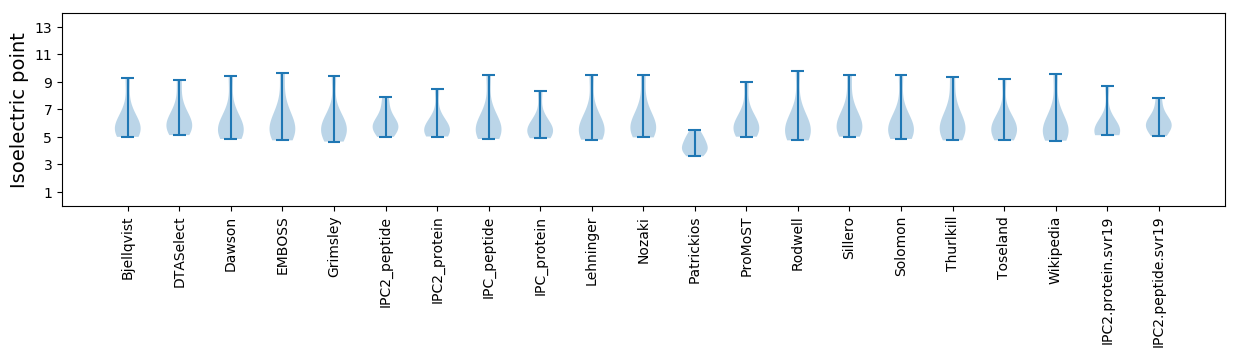
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L5YIM1|A0A1L5YIM1_9MONO Isoform of A0A1L5YIL7 W protein OS=Avian paramyxovirus 14 OX=1928005 GN=P PE=4 SV=1
MM1 pKa = 7.93DD2 pKa = 5.21FSDD5 pKa = 4.51DD6 pKa = 3.99NEE8 pKa = 4.13ITEE11 pKa = 4.89LLNISLDD18 pKa = 3.99TINKK22 pKa = 7.87IQHH25 pKa = 6.47AEE27 pKa = 4.15DD28 pKa = 3.71IKK30 pKa = 10.93PKK32 pKa = 9.93TVGKK36 pKa = 10.2SAIKK40 pKa = 10.05PGNTKK45 pKa = 10.64ALTKK49 pKa = 10.38AWEE52 pKa = 4.64DD53 pKa = 3.76DD54 pKa = 4.09SNTTSDD60 pKa = 3.07NTQNDD65 pKa = 3.08VRR67 pKa = 11.84GNSSAASNAQQSKK80 pKa = 10.21EE81 pKa = 3.88EE82 pKa = 4.38SGPEE86 pKa = 3.96TRR88 pKa = 11.84PEE90 pKa = 3.99TAGVEE95 pKa = 4.53VPSDD99 pKa = 3.76PTKK102 pKa = 10.82SSPDD106 pKa = 3.27QAEE109 pKa = 4.5TGWSRR114 pKa = 11.84GSNLDD119 pKa = 3.18KK120 pKa = 10.82TLEE123 pKa = 4.07NLEE126 pKa = 4.32KK127 pKa = 10.07ATSNRR132 pKa = 11.84VSDD135 pKa = 3.61QTQKK139 pKa = 10.51VLKK142 pKa = 8.93GGPVARR148 pKa = 11.84DD149 pKa = 3.14PLLRR153 pKa = 11.84RR154 pKa = 11.84PRR156 pKa = 3.81
MM1 pKa = 7.93DD2 pKa = 5.21FSDD5 pKa = 4.51DD6 pKa = 3.99NEE8 pKa = 4.13ITEE11 pKa = 4.89LLNISLDD18 pKa = 3.99TINKK22 pKa = 7.87IQHH25 pKa = 6.47AEE27 pKa = 4.15DD28 pKa = 3.71IKK30 pKa = 10.93PKK32 pKa = 9.93TVGKK36 pKa = 10.2SAIKK40 pKa = 10.05PGNTKK45 pKa = 10.64ALTKK49 pKa = 10.38AWEE52 pKa = 4.64DD53 pKa = 3.76DD54 pKa = 4.09SNTTSDD60 pKa = 3.07NTQNDD65 pKa = 3.08VRR67 pKa = 11.84GNSSAASNAQQSKK80 pKa = 10.21EE81 pKa = 3.88EE82 pKa = 4.38SGPEE86 pKa = 3.96TRR88 pKa = 11.84PEE90 pKa = 3.99TAGVEE95 pKa = 4.53VPSDD99 pKa = 3.76PTKK102 pKa = 10.82SSPDD106 pKa = 3.27QAEE109 pKa = 4.5TGWSRR114 pKa = 11.84GSNLDD119 pKa = 3.18KK120 pKa = 10.82TLEE123 pKa = 4.07NLEE126 pKa = 4.32KK127 pKa = 10.07ATSNRR132 pKa = 11.84VSDD135 pKa = 3.61QTQKK139 pKa = 10.51VLKK142 pKa = 8.93GGPVARR148 pKa = 11.84DD149 pKa = 3.14PLLRR153 pKa = 11.84RR154 pKa = 11.84PRR156 pKa = 3.81
Molecular weight: 16.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L5YIM9|A0A1L5YIM9_9MONO RNA-directed RNA polymerase L OS=Avian paramyxovirus 14 OX=1928005 GN=L PE=3 SV=1
MM1 pKa = 7.63ASASVNLFVDD11 pKa = 4.87EE12 pKa = 4.62SAPSSSLLAFPIVTKK27 pKa = 10.92DD28 pKa = 3.08NGSGGKK34 pKa = 9.57ALQPQIRR41 pKa = 11.84IAYY44 pKa = 8.16HH45 pKa = 6.4GEE47 pKa = 3.93VQGSKK52 pKa = 9.79RR53 pKa = 11.84SVMFVNCYY61 pKa = 10.45GFIEE65 pKa = 5.17DD66 pKa = 3.83YY67 pKa = 10.97SDD69 pKa = 3.78RR70 pKa = 11.84NEE72 pKa = 4.2GFLNPDD78 pKa = 3.64EE79 pKa = 4.61PPKK82 pKa = 10.58PRR84 pKa = 11.84TVTAACLTIGSVSGDD99 pKa = 3.39LDD101 pKa = 3.49PRR103 pKa = 11.84EE104 pKa = 4.07VARR107 pKa = 11.84ACFDD111 pKa = 4.09LDD113 pKa = 3.7VKK115 pKa = 11.06VKK117 pKa = 10.33INADD121 pKa = 3.04SRR123 pKa = 11.84EE124 pKa = 3.66RR125 pKa = 11.84VAFSFRR131 pKa = 11.84TKK133 pKa = 10.03PALLISSRR141 pKa = 11.84VISGGGIVLSAEE153 pKa = 4.33DD154 pKa = 4.03NIKK157 pKa = 10.74CVGKK161 pKa = 9.01MAVDD165 pKa = 3.03KK166 pKa = 10.57DD167 pKa = 3.48YY168 pKa = 11.22RR169 pKa = 11.84LRR171 pKa = 11.84IMFCSTTFLHH181 pKa = 6.39QSKK184 pKa = 10.18LFKK187 pKa = 10.57AQPAVMNLRR196 pKa = 11.84SSMLLALQVAVTIKK210 pKa = 10.38VDD212 pKa = 3.37IPASHH217 pKa = 7.11PHH219 pKa = 6.15AKK221 pKa = 10.07YY222 pKa = 10.38LSKK225 pKa = 11.23EE226 pKa = 3.84GDD228 pKa = 3.7DD229 pKa = 4.25LVAHH233 pKa = 6.07VWIHH237 pKa = 4.34VCNFKK242 pKa = 10.04RR243 pKa = 11.84TDD245 pKa = 3.1RR246 pKa = 11.84RR247 pKa = 11.84GEE249 pKa = 3.91TRR251 pKa = 11.84TVEE254 pKa = 3.92NLKK257 pKa = 10.57EE258 pKa = 4.1KK259 pKa = 9.69VRR261 pKa = 11.84RR262 pKa = 11.84MGLKK266 pKa = 10.33VSLLDD271 pKa = 3.29LWGPTIIIEE280 pKa = 4.34ATGTMTKK287 pKa = 10.22YY288 pKa = 11.05ALGFFSDD295 pKa = 4.57SKK297 pKa = 9.69TSCHH301 pKa = 6.82PISKK305 pKa = 10.19ASPEE309 pKa = 4.15TAKK312 pKa = 10.94LLWSATGEE320 pKa = 4.25VKK322 pKa = 10.57RR323 pKa = 11.84AVAVIQSSDD332 pKa = 2.88KK333 pKa = 10.75RR334 pKa = 11.84ALLTEE339 pKa = 5.04DD340 pKa = 3.92DD341 pKa = 4.77LLVKK345 pKa = 10.53GAMTTQAGGLKK356 pKa = 10.03EE357 pKa = 3.85FSLFKK362 pKa = 10.73KK363 pKa = 10.56
MM1 pKa = 7.63ASASVNLFVDD11 pKa = 4.87EE12 pKa = 4.62SAPSSSLLAFPIVTKK27 pKa = 10.92DD28 pKa = 3.08NGSGGKK34 pKa = 9.57ALQPQIRR41 pKa = 11.84IAYY44 pKa = 8.16HH45 pKa = 6.4GEE47 pKa = 3.93VQGSKK52 pKa = 9.79RR53 pKa = 11.84SVMFVNCYY61 pKa = 10.45GFIEE65 pKa = 5.17DD66 pKa = 3.83YY67 pKa = 10.97SDD69 pKa = 3.78RR70 pKa = 11.84NEE72 pKa = 4.2GFLNPDD78 pKa = 3.64EE79 pKa = 4.61PPKK82 pKa = 10.58PRR84 pKa = 11.84TVTAACLTIGSVSGDD99 pKa = 3.39LDD101 pKa = 3.49PRR103 pKa = 11.84EE104 pKa = 4.07VARR107 pKa = 11.84ACFDD111 pKa = 4.09LDD113 pKa = 3.7VKK115 pKa = 11.06VKK117 pKa = 10.33INADD121 pKa = 3.04SRR123 pKa = 11.84EE124 pKa = 3.66RR125 pKa = 11.84VAFSFRR131 pKa = 11.84TKK133 pKa = 10.03PALLISSRR141 pKa = 11.84VISGGGIVLSAEE153 pKa = 4.33DD154 pKa = 4.03NIKK157 pKa = 10.74CVGKK161 pKa = 9.01MAVDD165 pKa = 3.03KK166 pKa = 10.57DD167 pKa = 3.48YY168 pKa = 11.22RR169 pKa = 11.84LRR171 pKa = 11.84IMFCSTTFLHH181 pKa = 6.39QSKK184 pKa = 10.18LFKK187 pKa = 10.57AQPAVMNLRR196 pKa = 11.84SSMLLALQVAVTIKK210 pKa = 10.38VDD212 pKa = 3.37IPASHH217 pKa = 7.11PHH219 pKa = 6.15AKK221 pKa = 10.07YY222 pKa = 10.38LSKK225 pKa = 11.23EE226 pKa = 3.84GDD228 pKa = 3.7DD229 pKa = 4.25LVAHH233 pKa = 6.07VWIHH237 pKa = 4.34VCNFKK242 pKa = 10.04RR243 pKa = 11.84TDD245 pKa = 3.1RR246 pKa = 11.84RR247 pKa = 11.84GEE249 pKa = 3.91TRR251 pKa = 11.84TVEE254 pKa = 3.92NLKK257 pKa = 10.57EE258 pKa = 4.1KK259 pKa = 9.69VRR261 pKa = 11.84RR262 pKa = 11.84MGLKK266 pKa = 10.33VSLLDD271 pKa = 3.29LWGPTIIIEE280 pKa = 4.34ATGTMTKK287 pKa = 10.22YY288 pKa = 11.05ALGFFSDD295 pKa = 4.57SKK297 pKa = 9.69TSCHH301 pKa = 6.82PISKK305 pKa = 10.19ASPEE309 pKa = 4.15TAKK312 pKa = 10.94LLWSATGEE320 pKa = 4.25VKK322 pKa = 10.57RR323 pKa = 11.84AVAVIQSSDD332 pKa = 2.88KK333 pKa = 10.75RR334 pKa = 11.84ALLTEE339 pKa = 5.04DD340 pKa = 3.92DD341 pKa = 4.77LLVKK345 pKa = 10.53GAMTTQAGGLKK356 pKa = 10.03EE357 pKa = 3.85FSLFKK362 pKa = 10.73KK363 pKa = 10.56
Molecular weight: 39.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5006 |
156 |
2243 |
625.8 |
69.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.752 ± 0.655 | 2.058 ± 0.313 |
5.453 ± 0.382 | 5.733 ± 0.255 |
3.256 ± 0.422 | 6.312 ± 0.294 |
1.578 ± 0.227 | 7.231 ± 0.63 |
5.773 ± 0.351 | 9.409 ± 0.824 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.277 ± 0.323 | 4.914 ± 0.449 |
4.095 ± 0.408 | 4.055 ± 0.307 |
4.874 ± 0.409 | 9.189 ± 0.239 |
6.792 ± 0.592 | 6.153 ± 0.385 |
1.179 ± 0.231 | 2.917 ± 0.434 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
