
Avian metaavulavirus 11
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Metaavulavirus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
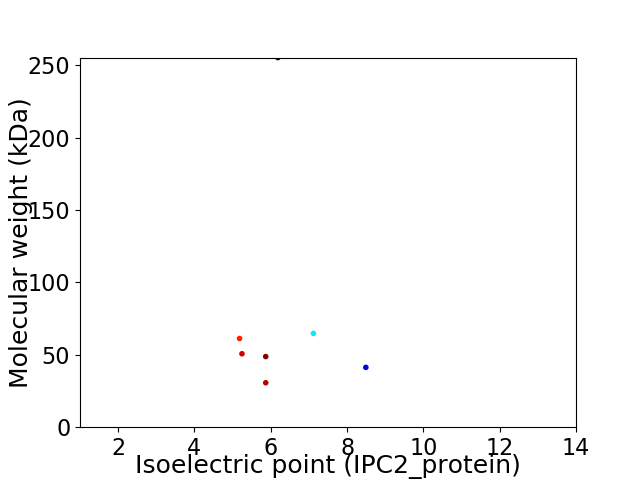
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|I6UTP1|I6UTP1_9MONO Matrix protein OS=Avian metaavulavirus 11 OX=2560310 PE=4 SV=1
MM1 pKa = 6.99GTCLNNRR8 pKa = 11.84LSTIPSIKK16 pKa = 9.42TVQCILIILSYY27 pKa = 10.14IIPYY31 pKa = 9.91SATDD35 pKa = 3.34NPIADD40 pKa = 4.15RR41 pKa = 11.84SLLRR45 pKa = 11.84AGIVPIYY52 pKa = 10.31SKK54 pKa = 11.02SLSVYY59 pKa = 8.18TNSISGYY66 pKa = 7.89LTVRR70 pKa = 11.84MLPPLPKK77 pKa = 10.47NLTEE81 pKa = 4.47CSQEE85 pKa = 4.08VVSNYY90 pKa = 10.36NKK92 pKa = 9.99TITRR96 pKa = 11.84MFQPISDD103 pKa = 3.49NLMRR107 pKa = 11.84IQEE110 pKa = 4.36GTDD113 pKa = 2.97SGTKK117 pKa = 9.68RR118 pKa = 11.84FVGAVIGSVALGVATSAQITAALAMVQAQDD148 pKa = 3.13NAKK151 pKa = 10.49AIWKK155 pKa = 9.12LKK157 pKa = 9.54EE158 pKa = 4.72AISSTNQAVLEE169 pKa = 4.25LKK171 pKa = 10.69EE172 pKa = 4.24GVNTLGVAVDD182 pKa = 4.72KK183 pKa = 11.03IQGYY187 pKa = 9.6INNEE191 pKa = 3.71ILPSLSEE198 pKa = 4.01LEE200 pKa = 4.38CRR202 pKa = 11.84VNANKK207 pKa = 10.11LASQLNLYY215 pKa = 10.13LIEE218 pKa = 4.33LTTIFGDD225 pKa = 4.07QITNPALTPLSLQALYY241 pKa = 9.87TLAGDD246 pKa = 3.79TMGSFLQYY254 pKa = 10.26IGAQDD259 pKa = 3.69NEE261 pKa = 4.34IEE263 pKa = 4.19SLYY266 pKa = 11.1DD267 pKa = 3.08SGLINGQIVSYY278 pKa = 10.07DD279 pKa = 3.37ASIQTIIIKK288 pKa = 10.22VSIPSISSLSRR299 pKa = 11.84FSIMRR304 pKa = 11.84LATVSSSVGGFEE316 pKa = 4.04KK317 pKa = 10.48TPLVPEE323 pKa = 4.08YY324 pKa = 11.25LLISDD329 pKa = 3.47NHH331 pKa = 7.44IEE333 pKa = 4.05EE334 pKa = 4.56FSIVDD339 pKa = 3.87CKK341 pKa = 10.73EE342 pKa = 3.81SSDD345 pKa = 4.44IFYY348 pKa = 10.58CPQILSMPISTATVEE363 pKa = 4.23CLKK366 pKa = 10.95GRR368 pKa = 11.84IDD370 pKa = 3.36QCIYY374 pKa = 10.36TSQLTILSHH383 pKa = 7.66RR384 pKa = 11.84IVTYY388 pKa = 10.57NGVVVANCFAEE399 pKa = 4.64LCRR402 pKa = 11.84CTNPSYY408 pKa = 10.72IIRR411 pKa = 11.84QDD413 pKa = 2.82RR414 pKa = 11.84DD415 pKa = 3.47VAVTVIDD422 pKa = 3.68KK423 pKa = 10.27DD424 pKa = 3.48LCKK427 pKa = 10.02RR428 pKa = 11.84VQIGDD433 pKa = 3.39IEE435 pKa = 5.53LIVQASIANEE445 pKa = 3.87YY446 pKa = 9.83KK447 pKa = 10.9VNFTVSEE454 pKa = 4.15DD455 pKa = 3.45QLAPSTPIDD464 pKa = 3.36ISNEE468 pKa = 3.91LNSLNQTLDD477 pKa = 3.18KK478 pKa = 10.94VGQLINTSNQILASLNPKK496 pKa = 9.48LVNNTSIIVLIVMGVVLILWLLALTIYY523 pKa = 10.47SIYY526 pKa = 10.59AARR529 pKa = 11.84NLNSIGRR536 pKa = 11.84LAKK539 pKa = 9.95SAYY542 pKa = 9.76ASYY545 pKa = 11.08VADD548 pKa = 3.41KK549 pKa = 10.96NVYY552 pKa = 9.6KK553 pKa = 10.93NEE555 pKa = 4.23STSSSSII562 pKa = 3.44
MM1 pKa = 6.99GTCLNNRR8 pKa = 11.84LSTIPSIKK16 pKa = 9.42TVQCILIILSYY27 pKa = 10.14IIPYY31 pKa = 9.91SATDD35 pKa = 3.34NPIADD40 pKa = 4.15RR41 pKa = 11.84SLLRR45 pKa = 11.84AGIVPIYY52 pKa = 10.31SKK54 pKa = 11.02SLSVYY59 pKa = 8.18TNSISGYY66 pKa = 7.89LTVRR70 pKa = 11.84MLPPLPKK77 pKa = 10.47NLTEE81 pKa = 4.47CSQEE85 pKa = 4.08VVSNYY90 pKa = 10.36NKK92 pKa = 9.99TITRR96 pKa = 11.84MFQPISDD103 pKa = 3.49NLMRR107 pKa = 11.84IQEE110 pKa = 4.36GTDD113 pKa = 2.97SGTKK117 pKa = 9.68RR118 pKa = 11.84FVGAVIGSVALGVATSAQITAALAMVQAQDD148 pKa = 3.13NAKK151 pKa = 10.49AIWKK155 pKa = 9.12LKK157 pKa = 9.54EE158 pKa = 4.72AISSTNQAVLEE169 pKa = 4.25LKK171 pKa = 10.69EE172 pKa = 4.24GVNTLGVAVDD182 pKa = 4.72KK183 pKa = 11.03IQGYY187 pKa = 9.6INNEE191 pKa = 3.71ILPSLSEE198 pKa = 4.01LEE200 pKa = 4.38CRR202 pKa = 11.84VNANKK207 pKa = 10.11LASQLNLYY215 pKa = 10.13LIEE218 pKa = 4.33LTTIFGDD225 pKa = 4.07QITNPALTPLSLQALYY241 pKa = 9.87TLAGDD246 pKa = 3.79TMGSFLQYY254 pKa = 10.26IGAQDD259 pKa = 3.69NEE261 pKa = 4.34IEE263 pKa = 4.19SLYY266 pKa = 11.1DD267 pKa = 3.08SGLINGQIVSYY278 pKa = 10.07DD279 pKa = 3.37ASIQTIIIKK288 pKa = 10.22VSIPSISSLSRR299 pKa = 11.84FSIMRR304 pKa = 11.84LATVSSSVGGFEE316 pKa = 4.04KK317 pKa = 10.48TPLVPEE323 pKa = 4.08YY324 pKa = 11.25LLISDD329 pKa = 3.47NHH331 pKa = 7.44IEE333 pKa = 4.05EE334 pKa = 4.56FSIVDD339 pKa = 3.87CKK341 pKa = 10.73EE342 pKa = 3.81SSDD345 pKa = 4.44IFYY348 pKa = 10.58CPQILSMPISTATVEE363 pKa = 4.23CLKK366 pKa = 10.95GRR368 pKa = 11.84IDD370 pKa = 3.36QCIYY374 pKa = 10.36TSQLTILSHH383 pKa = 7.66RR384 pKa = 11.84IVTYY388 pKa = 10.57NGVVVANCFAEE399 pKa = 4.64LCRR402 pKa = 11.84CTNPSYY408 pKa = 10.72IIRR411 pKa = 11.84QDD413 pKa = 2.82RR414 pKa = 11.84DD415 pKa = 3.47VAVTVIDD422 pKa = 3.68KK423 pKa = 10.27DD424 pKa = 3.48LCKK427 pKa = 10.02RR428 pKa = 11.84VQIGDD433 pKa = 3.39IEE435 pKa = 5.53LIVQASIANEE445 pKa = 3.87YY446 pKa = 9.83KK447 pKa = 10.9VNFTVSEE454 pKa = 4.15DD455 pKa = 3.45QLAPSTPIDD464 pKa = 3.36ISNEE468 pKa = 3.91LNSLNQTLDD477 pKa = 3.18KK478 pKa = 10.94VGQLINTSNQILASLNPKK496 pKa = 9.48LVNNTSIIVLIVMGVVLILWLLALTIYY523 pKa = 10.47SIYY526 pKa = 10.59AARR529 pKa = 11.84NLNSIGRR536 pKa = 11.84LAKK539 pKa = 9.95SAYY542 pKa = 9.76ASYY545 pKa = 11.08VADD548 pKa = 3.41KK549 pKa = 10.96NVYY552 pKa = 9.6KK553 pKa = 10.93NEE555 pKa = 4.23STSSSSII562 pKa = 3.44
Molecular weight: 61.28 kDa
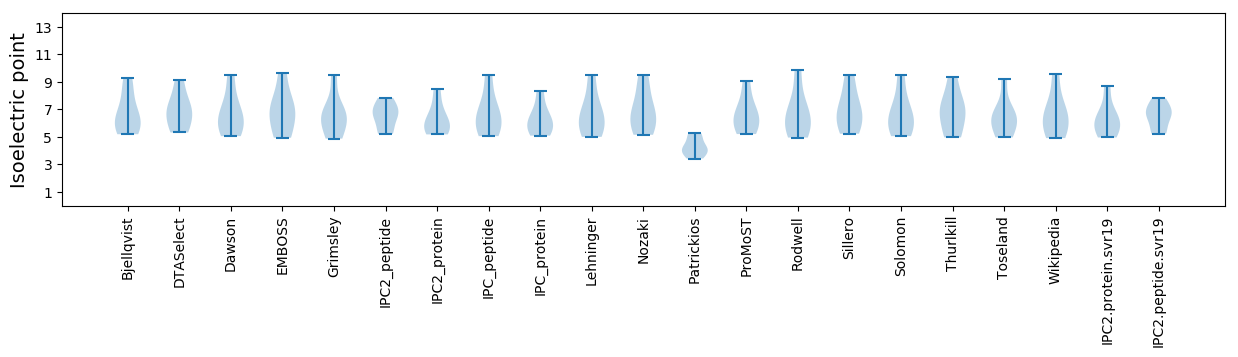
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I6UTP6|I6UTP6_9MONO Phosphoprotein OS=Avian metaavulavirus 11 OX=2560310 PE=4 SV=1
MM1 pKa = 7.27SLDD4 pKa = 3.69HH5 pKa = 6.44PTEE8 pKa = 4.83KK9 pKa = 10.39INLPIPTGHH18 pKa = 7.73PDD20 pKa = 3.48LEE22 pKa = 4.35LLAFPIIVEE31 pKa = 4.56RR32 pKa = 11.84GQDD35 pKa = 3.59GQKK38 pKa = 9.1TLQRR42 pKa = 11.84QYY44 pKa = 11.13RR45 pKa = 11.84IGSVGDD51 pKa = 3.24IFGSKK56 pKa = 7.67EE57 pKa = 3.63TGLFLTCYY65 pKa = 10.39GFIEE69 pKa = 4.37EE70 pKa = 5.54SYY72 pKa = 10.86TSTIQGKK79 pKa = 8.43SRR81 pKa = 11.84FFSEE85 pKa = 4.42HH86 pKa = 5.53QGGHH90 pKa = 5.9PKK92 pKa = 9.92IVTAALLPLGCVNPRR107 pKa = 11.84EE108 pKa = 4.56DD109 pKa = 3.62IDD111 pKa = 4.87RR112 pKa = 11.84LIEE115 pKa = 4.16DD116 pKa = 4.07SYY118 pKa = 9.71TLKK121 pKa = 11.02VLISKK126 pKa = 10.25NVDD129 pKa = 2.97SKK131 pKa = 11.31EE132 pKa = 3.8RR133 pKa = 11.84VVFKK137 pKa = 9.64FVNKK141 pKa = 9.24PAALSANKK149 pKa = 10.1LLINGGVILDD159 pKa = 3.9AEE161 pKa = 4.69AFVKK165 pKa = 10.68CPSKK169 pKa = 10.37LTTNMEE175 pKa = 3.68YY176 pKa = 10.3SFRR179 pKa = 11.84VVFVSVTRR187 pKa = 11.84LDD189 pKa = 3.27QTKK192 pKa = 10.06LYY194 pKa = 9.93RR195 pKa = 11.84VNEE198 pKa = 4.54SISKK202 pKa = 9.58IRR204 pKa = 11.84NPHH207 pKa = 6.53LIAVNLEE214 pKa = 4.2VKK216 pKa = 10.21ILLHH220 pKa = 6.02LTPDD224 pKa = 3.42NPQLKK229 pKa = 9.98FLIKK233 pKa = 10.05HH234 pKa = 5.53EE235 pKa = 4.41KK236 pKa = 10.34GGVGSVWIHH245 pKa = 6.17LCNFAKK251 pKa = 10.64KK252 pKa = 10.08NMKK255 pKa = 8.72GTLRR259 pKa = 11.84SIQDD263 pKa = 3.05VTEE266 pKa = 3.97KK267 pKa = 9.93VKK269 pKa = 10.9RR270 pKa = 11.84MALAVSLEE278 pKa = 4.25DD279 pKa = 3.1LWGPTVIVRR288 pKa = 11.84ANGTMSKK295 pKa = 10.2YY296 pKa = 10.93ALGFFSTSKK305 pKa = 9.04TACHH309 pKa = 7.04PIYY312 pKa = 10.7KK313 pKa = 9.94LAPEE317 pKa = 4.07IAKK320 pKa = 10.31IMWSCSSEE328 pKa = 3.77IIEE331 pKa = 5.13ANIIVQGSMKK341 pKa = 10.56NDD343 pKa = 2.97ILTGSDD349 pKa = 3.74LEE351 pKa = 4.44IPSTSKK357 pKa = 10.41IVNSGHH363 pKa = 5.73KK364 pKa = 10.13QFFSFKK370 pKa = 10.27KK371 pKa = 10.2
MM1 pKa = 7.27SLDD4 pKa = 3.69HH5 pKa = 6.44PTEE8 pKa = 4.83KK9 pKa = 10.39INLPIPTGHH18 pKa = 7.73PDD20 pKa = 3.48LEE22 pKa = 4.35LLAFPIIVEE31 pKa = 4.56RR32 pKa = 11.84GQDD35 pKa = 3.59GQKK38 pKa = 9.1TLQRR42 pKa = 11.84QYY44 pKa = 11.13RR45 pKa = 11.84IGSVGDD51 pKa = 3.24IFGSKK56 pKa = 7.67EE57 pKa = 3.63TGLFLTCYY65 pKa = 10.39GFIEE69 pKa = 4.37EE70 pKa = 5.54SYY72 pKa = 10.86TSTIQGKK79 pKa = 8.43SRR81 pKa = 11.84FFSEE85 pKa = 4.42HH86 pKa = 5.53QGGHH90 pKa = 5.9PKK92 pKa = 9.92IVTAALLPLGCVNPRR107 pKa = 11.84EE108 pKa = 4.56DD109 pKa = 3.62IDD111 pKa = 4.87RR112 pKa = 11.84LIEE115 pKa = 4.16DD116 pKa = 4.07SYY118 pKa = 9.71TLKK121 pKa = 11.02VLISKK126 pKa = 10.25NVDD129 pKa = 2.97SKK131 pKa = 11.31EE132 pKa = 3.8RR133 pKa = 11.84VVFKK137 pKa = 9.64FVNKK141 pKa = 9.24PAALSANKK149 pKa = 10.1LLINGGVILDD159 pKa = 3.9AEE161 pKa = 4.69AFVKK165 pKa = 10.68CPSKK169 pKa = 10.37LTTNMEE175 pKa = 3.68YY176 pKa = 10.3SFRR179 pKa = 11.84VVFVSVTRR187 pKa = 11.84LDD189 pKa = 3.27QTKK192 pKa = 10.06LYY194 pKa = 9.93RR195 pKa = 11.84VNEE198 pKa = 4.54SISKK202 pKa = 9.58IRR204 pKa = 11.84NPHH207 pKa = 6.53LIAVNLEE214 pKa = 4.2VKK216 pKa = 10.21ILLHH220 pKa = 6.02LTPDD224 pKa = 3.42NPQLKK229 pKa = 9.98FLIKK233 pKa = 10.05HH234 pKa = 5.53EE235 pKa = 4.41KK236 pKa = 10.34GGVGSVWIHH245 pKa = 6.17LCNFAKK251 pKa = 10.64KK252 pKa = 10.08NMKK255 pKa = 8.72GTLRR259 pKa = 11.84SIQDD263 pKa = 3.05VTEE266 pKa = 3.97KK267 pKa = 9.93VKK269 pKa = 10.9RR270 pKa = 11.84MALAVSLEE278 pKa = 4.25DD279 pKa = 3.1LWGPTVIVRR288 pKa = 11.84ANGTMSKK295 pKa = 10.2YY296 pKa = 10.93ALGFFSTSKK305 pKa = 9.04TACHH309 pKa = 7.04PIYY312 pKa = 10.7KK313 pKa = 9.94LAPEE317 pKa = 4.07IAKK320 pKa = 10.31IMWSCSSEE328 pKa = 3.77IIEE331 pKa = 5.13ANIIVQGSMKK341 pKa = 10.56NDD343 pKa = 2.97ILTGSDD349 pKa = 3.74LEE351 pKa = 4.44IPSTSKK357 pKa = 10.41IVNSGHH363 pKa = 5.73KK364 pKa = 10.13QFFSFKK370 pKa = 10.27KK371 pKa = 10.2
Molecular weight: 41.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4946 |
277 |
2251 |
706.6 |
79.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.803 ± 0.787 | 1.941 ± 0.294 |
5.156 ± 0.372 | 5.317 ± 0.324 |
3.255 ± 0.522 | 5.216 ± 0.383 |
2.082 ± 0.36 | 8.148 ± 0.69 |
4.873 ± 0.47 | 10.938 ± 0.949 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.123 ± 0.301 | 4.933 ± 0.323 |
4.792 ± 0.443 | 4.307 ± 0.396 |
5.176 ± 0.524 | 9.3 ± 0.597 |
6.692 ± 0.438 | 5.621 ± 0.472 |
1.072 ± 0.159 | 3.255 ± 0.385 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
