
Paludibacter jiangxiensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Paludibacteraceae; Paludibacter
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
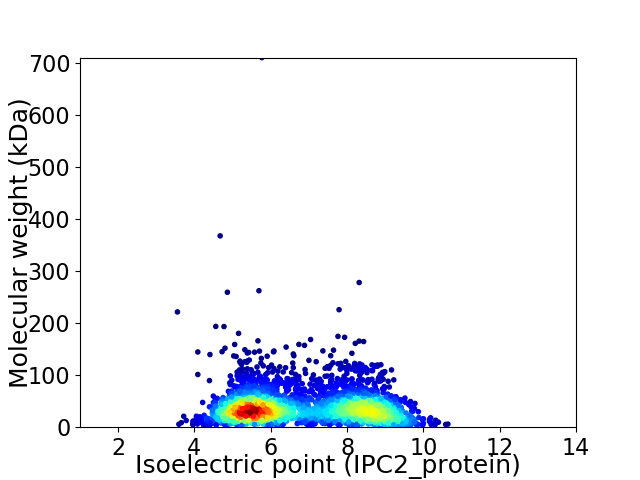
Virtual 2D-PAGE plot for 2945 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A171A247|A0A171A247_9BACT GxxExxY protein OS=Paludibacter jiangxiensis OX=681398 GN=PJIAN_3547 PE=4 SV=1
MM1 pKa = 7.41GGYY4 pKa = 10.38AGGLRR9 pKa = 11.84FFLLLAFICFEE20 pKa = 4.45SEE22 pKa = 4.02CSFNRR27 pKa = 11.84MFGQTHH33 pKa = 5.46VEE35 pKa = 3.95AGLSAVALKK44 pKa = 10.86QNTAFSFSPLSTRR57 pKa = 11.84RR58 pKa = 11.84TNVSEE63 pKa = 4.43ANHH66 pKa = 6.76DD67 pKa = 4.02PVASAASVTTLEE79 pKa = 4.11NTPVNGSVTATDD91 pKa = 3.45ADD93 pKa = 4.01GDD95 pKa = 3.96VLTFSKK101 pKa = 10.92VSDD104 pKa = 3.88PAHH107 pKa = 6.19GSVVVNADD115 pKa = 3.11GTFTYY120 pKa = 10.06TPYY123 pKa = 11.06LDD125 pKa = 4.13YY126 pKa = 11.16SGSDD130 pKa = 3.29SFVVGVTDD138 pKa = 4.28GNGGSATATVNVTVNHH154 pKa = 5.74VNQVPVITIRR164 pKa = 11.84QTTMEE169 pKa = 5.3DD170 pKa = 3.21SPIYY174 pKa = 10.45DD175 pKa = 3.54SVSASDD181 pKa = 4.75DD182 pKa = 4.16DD183 pKa = 4.75GDD185 pKa = 4.39NMTFSKK191 pKa = 10.77GISPLHH197 pKa = 6.24GALIVNSNGDD207 pKa = 3.66YY208 pKa = 10.85LYY210 pKa = 10.75TPDD213 pKa = 4.89KK214 pKa = 11.07DD215 pKa = 4.82FNGKK219 pKa = 9.92DD220 pKa = 3.18SFTITVNDD228 pKa = 3.93GNGGISTVKK237 pKa = 10.19IVVTVLPVNDD247 pKa = 4.91DD248 pKa = 3.8PVLTATPVTTQEE260 pKa = 4.0DD261 pKa = 4.86TPVNGSVTATDD272 pKa = 2.95VDD274 pKa = 4.21GDD276 pKa = 3.96VLTITKK282 pKa = 10.53DD283 pKa = 3.11SDD285 pKa = 3.87PLHH288 pKa = 5.82GTVVFNPDD296 pKa = 2.1GTYY299 pKa = 10.48AYY301 pKa = 9.23TPAQDD306 pKa = 3.99YY307 pKa = 11.32YY308 pKa = 11.24GDD310 pKa = 3.9DD311 pKa = 3.65SFTVSVSDD319 pKa = 4.15GNGGTATATVTITITSVNDD338 pKa = 3.45APVITATPVTTPEE351 pKa = 3.95DD352 pKa = 3.89TPVNGTATATDD363 pKa = 3.09VDD365 pKa = 4.23GDD367 pKa = 4.06VLTFALGTAPAHH379 pKa = 5.28GTAVVNSDD387 pKa = 2.74GSYY390 pKa = 10.39SYY392 pKa = 11.55SPAKK396 pKa = 10.3DD397 pKa = 3.6YY398 pKa = 11.76NGDD401 pKa = 3.56DD402 pKa = 3.56SFTIIVSDD410 pKa = 4.18GNGGTATATVTVTITPVNDD429 pKa = 3.85DD430 pKa = 4.05PVLTATPITTPEE442 pKa = 4.87DD443 pKa = 3.36IPVNGSVTATDD454 pKa = 2.95VDD456 pKa = 4.21GDD458 pKa = 3.96VLTITKK464 pKa = 10.42DD465 pKa = 2.95SDD467 pKa = 4.53PIHH470 pKa = 6.73GMVVFNPDD478 pKa = 2.24GTYY481 pKa = 10.11TYY483 pKa = 10.66TPAQDD488 pKa = 3.45YY489 pKa = 11.16HH490 pKa = 8.64GDD492 pKa = 3.54DD493 pKa = 3.78SFTVSVSDD501 pKa = 3.96GNGGTVTATVTVTITSVNDD520 pKa = 3.49NPVLTAPSVTTDD532 pKa = 2.9EE533 pKa = 4.43DD534 pKa = 4.11TSVNGSVTATDD545 pKa = 2.95VDD547 pKa = 4.21GDD549 pKa = 3.96VLTITKK555 pKa = 10.53DD556 pKa = 3.11SDD558 pKa = 3.87PLHH561 pKa = 5.78GTVVFNSDD569 pKa = 2.46GTYY572 pKa = 10.08TYY574 pKa = 10.77TPAQDD579 pKa = 3.35YY580 pKa = 10.93HH581 pKa = 7.47GADD584 pKa = 3.16SFTVNVSDD592 pKa = 4.84GNGGTATGTVSITVISVNDD611 pKa = 3.77DD612 pKa = 3.26PVITVTPVTTQEE624 pKa = 3.92DD625 pKa = 4.82TPVNGTATATDD636 pKa = 3.37VDD638 pKa = 4.15GDD640 pKa = 3.98ALTFSKK646 pKa = 10.9GSDD649 pKa = 3.57PSHH652 pKa = 5.89GTVDD656 pKa = 3.32VHH658 pKa = 8.84SDD660 pKa = 2.88GTYY663 pKa = 9.71TYY665 pKa = 10.66TPVKK669 pKa = 10.17DD670 pKa = 3.87YY671 pKa = 11.66NGDD674 pKa = 3.52DD675 pKa = 3.62SFTISVSDD683 pKa = 3.75GNGGVASATVTVTITPVNDD702 pKa = 3.4APVLSDD708 pKa = 3.57TPVTTQEE715 pKa = 3.9DD716 pKa = 4.75TPLNGSVTATDD727 pKa = 3.24VDD729 pKa = 4.31GDD731 pKa = 3.89ALTYY735 pKa = 11.04SKK737 pKa = 11.15DD738 pKa = 3.3SDD740 pKa = 4.02PSHH743 pKa = 6.04GTLVFNSDD751 pKa = 2.64GTYY754 pKa = 10.08TYY756 pKa = 10.77TPAQDD761 pKa = 3.35YY762 pKa = 10.93HH763 pKa = 7.47GADD766 pKa = 3.33SFTVSVSDD774 pKa = 4.15GNGGTATGTVSITVISVNDD793 pKa = 3.77DD794 pKa = 3.26PVITVTPVTTPEE806 pKa = 3.89DD807 pKa = 3.89TPVNGTATATDD818 pKa = 3.09VDD820 pKa = 4.23GDD822 pKa = 4.02VLTFAVGTAPSHH834 pKa = 5.51GTVDD838 pKa = 3.34VHH840 pKa = 8.84SDD842 pKa = 2.88GTYY845 pKa = 9.71TYY847 pKa = 10.66TPVKK851 pKa = 10.17DD852 pKa = 3.87YY853 pKa = 11.66NGDD856 pKa = 3.52DD857 pKa = 3.62SFTISVSDD865 pKa = 3.75GNGGVASATVTVTITPVNDD884 pKa = 3.4APVLSDD890 pKa = 3.57TPVTTQEE897 pKa = 3.9DD898 pKa = 4.75TPLNGSVTATDD909 pKa = 3.24VDD911 pKa = 4.31GDD913 pKa = 3.89ALTYY917 pKa = 11.04SKK919 pKa = 11.15DD920 pKa = 3.3SDD922 pKa = 4.02PSHH925 pKa = 6.04GTLVFNSDD933 pKa = 2.64GTYY936 pKa = 9.94TYY938 pKa = 10.77TPATDD943 pKa = 3.4YY944 pKa = 11.41HH945 pKa = 8.13GDD947 pKa = 3.47DD948 pKa = 3.86SFTVSVSDD956 pKa = 4.15GNGGTATGTVSITVISVNDD975 pKa = 3.77DD976 pKa = 3.26PVITVTPVTTQEE988 pKa = 4.05DD989 pKa = 4.97TPVSGTVTATDD1000 pKa = 3.16VDD1002 pKa = 4.04GDD1004 pKa = 3.98ALTFSKK1010 pKa = 10.9GSDD1013 pKa = 3.57PSHH1016 pKa = 5.89GTVDD1020 pKa = 3.38VHH1022 pKa = 8.73SDD1024 pKa = 2.87GSYY1027 pKa = 9.93TYY1029 pKa = 10.56TPAKK1033 pKa = 10.1DD1034 pKa = 3.7YY1035 pKa = 11.66NGDD1038 pKa = 3.61DD1039 pKa = 3.31SFMISVSDD1047 pKa = 4.05GNGGTATGTVTVTITPVNDD1066 pKa = 3.95DD1067 pKa = 3.9PVLAATLVTTQEE1079 pKa = 4.93DD1080 pKa = 4.03IPLNASVIATDD1091 pKa = 3.27VDD1093 pKa = 4.06GDD1095 pKa = 3.88ALTYY1099 pKa = 11.04SKK1101 pKa = 11.15DD1102 pKa = 3.3SDD1104 pKa = 4.02PSHH1107 pKa = 6.04GTLVFNSDD1115 pKa = 2.64GTYY1118 pKa = 9.94TYY1120 pKa = 10.77TPATDD1125 pKa = 3.4YY1126 pKa = 11.41HH1127 pKa = 8.13GDD1129 pKa = 3.47DD1130 pKa = 3.86SFTVSVSDD1138 pKa = 4.15GNGGTATGTVSITVISVNDD1157 pKa = 3.77DD1158 pKa = 3.26PVITVTPVTTQEE1170 pKa = 4.05DD1171 pKa = 4.95TPVSGTATATDD1182 pKa = 3.37VDD1184 pKa = 4.15GDD1186 pKa = 3.98ALTFSKK1192 pKa = 10.9GSDD1195 pKa = 3.57PSHH1198 pKa = 5.89GTVDD1202 pKa = 3.38VHH1204 pKa = 8.73SDD1206 pKa = 2.87GSYY1209 pKa = 9.93TYY1211 pKa = 10.56TPAKK1215 pKa = 10.14DD1216 pKa = 3.66YY1217 pKa = 11.67NGDD1220 pKa = 3.57DD1221 pKa = 3.33TFMISVSDD1229 pKa = 3.9GNGGVASGTVTVTITPVNDD1248 pKa = 3.89DD1249 pKa = 3.93PVTTLLSVSTPEE1261 pKa = 3.93DD1262 pKa = 3.77TPLVSTVMAGATDD1275 pKa = 3.19VDD1277 pKa = 4.2GDD1279 pKa = 4.02VLTVTGFTMDD1289 pKa = 3.29GTNYY1293 pKa = 10.33LPGTAVVISGKK1304 pKa = 8.51GTFILNANGDD1314 pKa = 3.89FTLTPDD1320 pKa = 3.44KK1321 pKa = 10.5DD1322 pKa = 3.97YY1323 pKa = 11.63NGAVPSVTYY1332 pKa = 9.37TVVDD1336 pKa = 4.29GNGGTAVGTLNLTVIAVNDD1355 pKa = 3.93DD1356 pKa = 3.41PVITVTPVTTQEE1368 pKa = 3.92DD1369 pKa = 4.82TPVNGTATATDD1380 pKa = 3.37VDD1382 pKa = 4.15GDD1384 pKa = 3.98ALTFSKK1390 pKa = 10.91GSDD1393 pKa = 3.57PVHH1396 pKa = 5.81GTVDD1400 pKa = 3.34VHH1402 pKa = 8.59SDD1404 pKa = 2.87GSYY1407 pKa = 9.93TYY1409 pKa = 10.56TPAKK1413 pKa = 10.25DD1414 pKa = 3.6YY1415 pKa = 11.66NGDD1418 pKa = 3.76DD1419 pKa = 3.6NFTISVSDD1427 pKa = 3.94GNGGTASATVTVTITPVNDD1446 pKa = 3.48APVLIATPVTTQEE1459 pKa = 4.88DD1460 pKa = 4.04IPLNGSVTATDD1471 pKa = 3.24VDD1473 pKa = 3.99GDD1475 pKa = 4.0ALTFSKK1481 pKa = 10.9DD1482 pKa = 2.99SDD1484 pKa = 4.02PSHH1487 pKa = 6.04GTLVFNSDD1495 pKa = 2.64GTYY1498 pKa = 9.94TYY1500 pKa = 10.77TPATDD1505 pKa = 3.4YY1506 pKa = 11.41HH1507 pKa = 8.13GDD1509 pKa = 3.47DD1510 pKa = 3.86SFTVSVSDD1518 pKa = 4.15GNGGTATGTVSITVISVNDD1537 pKa = 3.77DD1538 pKa = 3.26PVITVTPVTTQEE1550 pKa = 3.99DD1551 pKa = 4.36TSVNGTATATDD1562 pKa = 3.37VDD1564 pKa = 4.15GDD1566 pKa = 3.98ALTFSKK1572 pKa = 10.9GSDD1575 pKa = 3.57PSHH1578 pKa = 5.89GTVDD1582 pKa = 3.38VHH1584 pKa = 8.72SDD1586 pKa = 2.8GSYY1589 pKa = 10.85VYY1591 pKa = 10.38TPAKK1595 pKa = 10.28DD1596 pKa = 3.61YY1597 pKa = 11.69NGDD1600 pKa = 3.76DD1601 pKa = 3.6NFTISVSDD1609 pKa = 3.92GNGGAASATVTVIITPVNDD1628 pKa = 3.73APVLTATPVTTQEE1641 pKa = 5.03DD1642 pKa = 3.92IPANGNVTATDD1653 pKa = 3.23VDD1655 pKa = 3.71GDD1657 pKa = 4.06ALIFTKK1663 pKa = 10.67DD1664 pKa = 3.21SDD1666 pKa = 4.28PLHH1669 pKa = 5.78GTVVFNTDD1677 pKa = 2.17GTYY1680 pKa = 10.06TYY1682 pKa = 10.7TPATDD1687 pKa = 3.34YY1688 pKa = 11.25HH1689 pKa = 6.39GADD1692 pKa = 3.11SFMVSVSDD1700 pKa = 4.31GNGGTATATVLITIVSVNDD1719 pKa = 3.63DD1720 pKa = 3.52PVITVTPVTTQEE1732 pKa = 4.16DD1733 pKa = 4.4TSVTGTATATDD1744 pKa = 3.15VDD1746 pKa = 4.11GDD1748 pKa = 4.04VLSFAVGIASSHH1760 pKa = 5.11GTVTVNPDD1768 pKa = 2.67GTYY1771 pKa = 10.09TYY1773 pKa = 10.68IPAKK1777 pKa = 10.16DD1778 pKa = 3.68YY1779 pKa = 11.53NGNDD1783 pKa = 3.3SFTVTVSDD1791 pKa = 4.43GNGGTATATVTVTITSVNDD1810 pKa = 3.06VPVAKK1815 pKa = 10.07DD1816 pKa = 3.31DD1817 pKa = 3.8AVSVVEE1823 pKa = 4.3CTSVSGTVAGNDD1835 pKa = 3.65TPSGDD1840 pKa = 3.76GGNVWSVNKK1849 pKa = 10.08NPSHH1853 pKa = 6.02GTVTLNADD1861 pKa = 3.14GTYY1864 pKa = 9.91IYY1866 pKa = 10.32IPTPNYY1872 pKa = 9.9HH1873 pKa = 5.7GTDD1876 pKa = 3.16TFVYY1880 pKa = 9.92TLTDD1884 pKa = 2.98VDD1886 pKa = 4.63GDD1888 pKa = 3.62QSTATMSITVTPLSEE1903 pKa = 3.88KK1904 pKa = 10.8LLIIKK1909 pKa = 10.2QSTKK1913 pKa = 10.1PQQTSDD1919 pKa = 3.11GSFIWKK1925 pKa = 8.11YY1926 pKa = 9.48TISLTNLQNIIVDD1939 pKa = 4.42NIQVEE1944 pKa = 4.32DD1945 pKa = 4.27DD1946 pKa = 3.49LSKK1949 pKa = 11.24VFTHH1953 pKa = 6.69GEE1955 pKa = 4.13SFKK1958 pKa = 10.12VTNITASGNLKK1969 pKa = 10.95ANGLYY1974 pKa = 10.6DD1975 pKa = 3.86GLNSMSTLLSGSYY1988 pKa = 9.8LAPLSKK1994 pKa = 10.69DD1995 pKa = 3.45SIVIEE2000 pKa = 4.2VQVYY2004 pKa = 7.27SHH2006 pKa = 6.37NFIGNVYY2013 pKa = 9.68NQALFEE2019 pKa = 4.43GFSNSTGQIKK2029 pKa = 10.1DD2030 pKa = 3.61AQSDD2034 pKa = 3.95DD2035 pKa = 3.99PLNTEE2040 pKa = 5.1ASPSSPRR2047 pKa = 11.84TTITFIPEE2055 pKa = 4.01VEE2057 pKa = 3.97LHH2059 pKa = 5.88IPEE2062 pKa = 4.68GFSPNKK2068 pKa = 10.35DD2069 pKa = 3.16MYY2071 pKa = 11.03NDD2073 pKa = 3.39TFEE2076 pKa = 4.57IVHH2079 pKa = 6.3SGNVTLEE2086 pKa = 3.9VEE2088 pKa = 4.55VFNRR2092 pKa = 11.84WGNRR2096 pKa = 11.84VYY2098 pKa = 10.93RR2099 pKa = 11.84STDD2102 pKa = 3.18YY2103 pKa = 11.42HH2104 pKa = 7.34DD2105 pKa = 5.03DD2106 pKa = 3.56WDD2108 pKa = 4.78GKK2110 pKa = 11.06GADD2113 pKa = 3.75NLLGKK2118 pKa = 9.77DD2119 pKa = 4.06LPEE2122 pKa = 3.81GTYY2125 pKa = 10.11FYY2127 pKa = 10.73IVTTTNKK2134 pKa = 8.57KK2135 pKa = 7.56TSEE2138 pKa = 3.98VKK2140 pKa = 10.3KK2141 pKa = 10.44IKK2143 pKa = 10.64GSVTLRR2149 pKa = 11.84RR2150 pKa = 3.8
MM1 pKa = 7.41GGYY4 pKa = 10.38AGGLRR9 pKa = 11.84FFLLLAFICFEE20 pKa = 4.45SEE22 pKa = 4.02CSFNRR27 pKa = 11.84MFGQTHH33 pKa = 5.46VEE35 pKa = 3.95AGLSAVALKK44 pKa = 10.86QNTAFSFSPLSTRR57 pKa = 11.84RR58 pKa = 11.84TNVSEE63 pKa = 4.43ANHH66 pKa = 6.76DD67 pKa = 4.02PVASAASVTTLEE79 pKa = 4.11NTPVNGSVTATDD91 pKa = 3.45ADD93 pKa = 4.01GDD95 pKa = 3.96VLTFSKK101 pKa = 10.92VSDD104 pKa = 3.88PAHH107 pKa = 6.19GSVVVNADD115 pKa = 3.11GTFTYY120 pKa = 10.06TPYY123 pKa = 11.06LDD125 pKa = 4.13YY126 pKa = 11.16SGSDD130 pKa = 3.29SFVVGVTDD138 pKa = 4.28GNGGSATATVNVTVNHH154 pKa = 5.74VNQVPVITIRR164 pKa = 11.84QTTMEE169 pKa = 5.3DD170 pKa = 3.21SPIYY174 pKa = 10.45DD175 pKa = 3.54SVSASDD181 pKa = 4.75DD182 pKa = 4.16DD183 pKa = 4.75GDD185 pKa = 4.39NMTFSKK191 pKa = 10.77GISPLHH197 pKa = 6.24GALIVNSNGDD207 pKa = 3.66YY208 pKa = 10.85LYY210 pKa = 10.75TPDD213 pKa = 4.89KK214 pKa = 11.07DD215 pKa = 4.82FNGKK219 pKa = 9.92DD220 pKa = 3.18SFTITVNDD228 pKa = 3.93GNGGISTVKK237 pKa = 10.19IVVTVLPVNDD247 pKa = 4.91DD248 pKa = 3.8PVLTATPVTTQEE260 pKa = 4.0DD261 pKa = 4.86TPVNGSVTATDD272 pKa = 2.95VDD274 pKa = 4.21GDD276 pKa = 3.96VLTITKK282 pKa = 10.53DD283 pKa = 3.11SDD285 pKa = 3.87PLHH288 pKa = 5.82GTVVFNPDD296 pKa = 2.1GTYY299 pKa = 10.48AYY301 pKa = 9.23TPAQDD306 pKa = 3.99YY307 pKa = 11.32YY308 pKa = 11.24GDD310 pKa = 3.9DD311 pKa = 3.65SFTVSVSDD319 pKa = 4.15GNGGTATATVTITITSVNDD338 pKa = 3.45APVITATPVTTPEE351 pKa = 3.95DD352 pKa = 3.89TPVNGTATATDD363 pKa = 3.09VDD365 pKa = 4.23GDD367 pKa = 4.06VLTFALGTAPAHH379 pKa = 5.28GTAVVNSDD387 pKa = 2.74GSYY390 pKa = 10.39SYY392 pKa = 11.55SPAKK396 pKa = 10.3DD397 pKa = 3.6YY398 pKa = 11.76NGDD401 pKa = 3.56DD402 pKa = 3.56SFTIIVSDD410 pKa = 4.18GNGGTATATVTVTITPVNDD429 pKa = 3.85DD430 pKa = 4.05PVLTATPITTPEE442 pKa = 4.87DD443 pKa = 3.36IPVNGSVTATDD454 pKa = 2.95VDD456 pKa = 4.21GDD458 pKa = 3.96VLTITKK464 pKa = 10.42DD465 pKa = 2.95SDD467 pKa = 4.53PIHH470 pKa = 6.73GMVVFNPDD478 pKa = 2.24GTYY481 pKa = 10.11TYY483 pKa = 10.66TPAQDD488 pKa = 3.45YY489 pKa = 11.16HH490 pKa = 8.64GDD492 pKa = 3.54DD493 pKa = 3.78SFTVSVSDD501 pKa = 3.96GNGGTVTATVTVTITSVNDD520 pKa = 3.49NPVLTAPSVTTDD532 pKa = 2.9EE533 pKa = 4.43DD534 pKa = 4.11TSVNGSVTATDD545 pKa = 2.95VDD547 pKa = 4.21GDD549 pKa = 3.96VLTITKK555 pKa = 10.53DD556 pKa = 3.11SDD558 pKa = 3.87PLHH561 pKa = 5.78GTVVFNSDD569 pKa = 2.46GTYY572 pKa = 10.08TYY574 pKa = 10.77TPAQDD579 pKa = 3.35YY580 pKa = 10.93HH581 pKa = 7.47GADD584 pKa = 3.16SFTVNVSDD592 pKa = 4.84GNGGTATGTVSITVISVNDD611 pKa = 3.77DD612 pKa = 3.26PVITVTPVTTQEE624 pKa = 3.92DD625 pKa = 4.82TPVNGTATATDD636 pKa = 3.37VDD638 pKa = 4.15GDD640 pKa = 3.98ALTFSKK646 pKa = 10.9GSDD649 pKa = 3.57PSHH652 pKa = 5.89GTVDD656 pKa = 3.32VHH658 pKa = 8.84SDD660 pKa = 2.88GTYY663 pKa = 9.71TYY665 pKa = 10.66TPVKK669 pKa = 10.17DD670 pKa = 3.87YY671 pKa = 11.66NGDD674 pKa = 3.52DD675 pKa = 3.62SFTISVSDD683 pKa = 3.75GNGGVASATVTVTITPVNDD702 pKa = 3.4APVLSDD708 pKa = 3.57TPVTTQEE715 pKa = 3.9DD716 pKa = 4.75TPLNGSVTATDD727 pKa = 3.24VDD729 pKa = 4.31GDD731 pKa = 3.89ALTYY735 pKa = 11.04SKK737 pKa = 11.15DD738 pKa = 3.3SDD740 pKa = 4.02PSHH743 pKa = 6.04GTLVFNSDD751 pKa = 2.64GTYY754 pKa = 10.08TYY756 pKa = 10.77TPAQDD761 pKa = 3.35YY762 pKa = 10.93HH763 pKa = 7.47GADD766 pKa = 3.33SFTVSVSDD774 pKa = 4.15GNGGTATGTVSITVISVNDD793 pKa = 3.77DD794 pKa = 3.26PVITVTPVTTPEE806 pKa = 3.89DD807 pKa = 3.89TPVNGTATATDD818 pKa = 3.09VDD820 pKa = 4.23GDD822 pKa = 4.02VLTFAVGTAPSHH834 pKa = 5.51GTVDD838 pKa = 3.34VHH840 pKa = 8.84SDD842 pKa = 2.88GTYY845 pKa = 9.71TYY847 pKa = 10.66TPVKK851 pKa = 10.17DD852 pKa = 3.87YY853 pKa = 11.66NGDD856 pKa = 3.52DD857 pKa = 3.62SFTISVSDD865 pKa = 3.75GNGGVASATVTVTITPVNDD884 pKa = 3.4APVLSDD890 pKa = 3.57TPVTTQEE897 pKa = 3.9DD898 pKa = 4.75TPLNGSVTATDD909 pKa = 3.24VDD911 pKa = 4.31GDD913 pKa = 3.89ALTYY917 pKa = 11.04SKK919 pKa = 11.15DD920 pKa = 3.3SDD922 pKa = 4.02PSHH925 pKa = 6.04GTLVFNSDD933 pKa = 2.64GTYY936 pKa = 9.94TYY938 pKa = 10.77TPATDD943 pKa = 3.4YY944 pKa = 11.41HH945 pKa = 8.13GDD947 pKa = 3.47DD948 pKa = 3.86SFTVSVSDD956 pKa = 4.15GNGGTATGTVSITVISVNDD975 pKa = 3.77DD976 pKa = 3.26PVITVTPVTTQEE988 pKa = 4.05DD989 pKa = 4.97TPVSGTVTATDD1000 pKa = 3.16VDD1002 pKa = 4.04GDD1004 pKa = 3.98ALTFSKK1010 pKa = 10.9GSDD1013 pKa = 3.57PSHH1016 pKa = 5.89GTVDD1020 pKa = 3.38VHH1022 pKa = 8.73SDD1024 pKa = 2.87GSYY1027 pKa = 9.93TYY1029 pKa = 10.56TPAKK1033 pKa = 10.1DD1034 pKa = 3.7YY1035 pKa = 11.66NGDD1038 pKa = 3.61DD1039 pKa = 3.31SFMISVSDD1047 pKa = 4.05GNGGTATGTVTVTITPVNDD1066 pKa = 3.95DD1067 pKa = 3.9PVLAATLVTTQEE1079 pKa = 4.93DD1080 pKa = 4.03IPLNASVIATDD1091 pKa = 3.27VDD1093 pKa = 4.06GDD1095 pKa = 3.88ALTYY1099 pKa = 11.04SKK1101 pKa = 11.15DD1102 pKa = 3.3SDD1104 pKa = 4.02PSHH1107 pKa = 6.04GTLVFNSDD1115 pKa = 2.64GTYY1118 pKa = 9.94TYY1120 pKa = 10.77TPATDD1125 pKa = 3.4YY1126 pKa = 11.41HH1127 pKa = 8.13GDD1129 pKa = 3.47DD1130 pKa = 3.86SFTVSVSDD1138 pKa = 4.15GNGGTATGTVSITVISVNDD1157 pKa = 3.77DD1158 pKa = 3.26PVITVTPVTTQEE1170 pKa = 4.05DD1171 pKa = 4.95TPVSGTATATDD1182 pKa = 3.37VDD1184 pKa = 4.15GDD1186 pKa = 3.98ALTFSKK1192 pKa = 10.9GSDD1195 pKa = 3.57PSHH1198 pKa = 5.89GTVDD1202 pKa = 3.38VHH1204 pKa = 8.73SDD1206 pKa = 2.87GSYY1209 pKa = 9.93TYY1211 pKa = 10.56TPAKK1215 pKa = 10.14DD1216 pKa = 3.66YY1217 pKa = 11.67NGDD1220 pKa = 3.57DD1221 pKa = 3.33TFMISVSDD1229 pKa = 3.9GNGGVASGTVTVTITPVNDD1248 pKa = 3.89DD1249 pKa = 3.93PVTTLLSVSTPEE1261 pKa = 3.93DD1262 pKa = 3.77TPLVSTVMAGATDD1275 pKa = 3.19VDD1277 pKa = 4.2GDD1279 pKa = 4.02VLTVTGFTMDD1289 pKa = 3.29GTNYY1293 pKa = 10.33LPGTAVVISGKK1304 pKa = 8.51GTFILNANGDD1314 pKa = 3.89FTLTPDD1320 pKa = 3.44KK1321 pKa = 10.5DD1322 pKa = 3.97YY1323 pKa = 11.63NGAVPSVTYY1332 pKa = 9.37TVVDD1336 pKa = 4.29GNGGTAVGTLNLTVIAVNDD1355 pKa = 3.93DD1356 pKa = 3.41PVITVTPVTTQEE1368 pKa = 3.92DD1369 pKa = 4.82TPVNGTATATDD1380 pKa = 3.37VDD1382 pKa = 4.15GDD1384 pKa = 3.98ALTFSKK1390 pKa = 10.91GSDD1393 pKa = 3.57PVHH1396 pKa = 5.81GTVDD1400 pKa = 3.34VHH1402 pKa = 8.59SDD1404 pKa = 2.87GSYY1407 pKa = 9.93TYY1409 pKa = 10.56TPAKK1413 pKa = 10.25DD1414 pKa = 3.6YY1415 pKa = 11.66NGDD1418 pKa = 3.76DD1419 pKa = 3.6NFTISVSDD1427 pKa = 3.94GNGGTASATVTVTITPVNDD1446 pKa = 3.48APVLIATPVTTQEE1459 pKa = 4.88DD1460 pKa = 4.04IPLNGSVTATDD1471 pKa = 3.24VDD1473 pKa = 3.99GDD1475 pKa = 4.0ALTFSKK1481 pKa = 10.9DD1482 pKa = 2.99SDD1484 pKa = 4.02PSHH1487 pKa = 6.04GTLVFNSDD1495 pKa = 2.64GTYY1498 pKa = 9.94TYY1500 pKa = 10.77TPATDD1505 pKa = 3.4YY1506 pKa = 11.41HH1507 pKa = 8.13GDD1509 pKa = 3.47DD1510 pKa = 3.86SFTVSVSDD1518 pKa = 4.15GNGGTATGTVSITVISVNDD1537 pKa = 3.77DD1538 pKa = 3.26PVITVTPVTTQEE1550 pKa = 3.99DD1551 pKa = 4.36TSVNGTATATDD1562 pKa = 3.37VDD1564 pKa = 4.15GDD1566 pKa = 3.98ALTFSKK1572 pKa = 10.9GSDD1575 pKa = 3.57PSHH1578 pKa = 5.89GTVDD1582 pKa = 3.38VHH1584 pKa = 8.72SDD1586 pKa = 2.8GSYY1589 pKa = 10.85VYY1591 pKa = 10.38TPAKK1595 pKa = 10.28DD1596 pKa = 3.61YY1597 pKa = 11.69NGDD1600 pKa = 3.76DD1601 pKa = 3.6NFTISVSDD1609 pKa = 3.92GNGGAASATVTVIITPVNDD1628 pKa = 3.73APVLTATPVTTQEE1641 pKa = 5.03DD1642 pKa = 3.92IPANGNVTATDD1653 pKa = 3.23VDD1655 pKa = 3.71GDD1657 pKa = 4.06ALIFTKK1663 pKa = 10.67DD1664 pKa = 3.21SDD1666 pKa = 4.28PLHH1669 pKa = 5.78GTVVFNTDD1677 pKa = 2.17GTYY1680 pKa = 10.06TYY1682 pKa = 10.7TPATDD1687 pKa = 3.34YY1688 pKa = 11.25HH1689 pKa = 6.39GADD1692 pKa = 3.11SFMVSVSDD1700 pKa = 4.31GNGGTATATVLITIVSVNDD1719 pKa = 3.63DD1720 pKa = 3.52PVITVTPVTTQEE1732 pKa = 4.16DD1733 pKa = 4.4TSVTGTATATDD1744 pKa = 3.15VDD1746 pKa = 4.11GDD1748 pKa = 4.04VLSFAVGIASSHH1760 pKa = 5.11GTVTVNPDD1768 pKa = 2.67GTYY1771 pKa = 10.09TYY1773 pKa = 10.68IPAKK1777 pKa = 10.16DD1778 pKa = 3.68YY1779 pKa = 11.53NGNDD1783 pKa = 3.3SFTVTVSDD1791 pKa = 4.43GNGGTATATVTVTITSVNDD1810 pKa = 3.06VPVAKK1815 pKa = 10.07DD1816 pKa = 3.31DD1817 pKa = 3.8AVSVVEE1823 pKa = 4.3CTSVSGTVAGNDD1835 pKa = 3.65TPSGDD1840 pKa = 3.76GGNVWSVNKK1849 pKa = 10.08NPSHH1853 pKa = 6.02GTVTLNADD1861 pKa = 3.14GTYY1864 pKa = 9.91IYY1866 pKa = 10.32IPTPNYY1872 pKa = 9.9HH1873 pKa = 5.7GTDD1876 pKa = 3.16TFVYY1880 pKa = 9.92TLTDD1884 pKa = 2.98VDD1886 pKa = 4.63GDD1888 pKa = 3.62QSTATMSITVTPLSEE1903 pKa = 3.88KK1904 pKa = 10.8LLIIKK1909 pKa = 10.2QSTKK1913 pKa = 10.1PQQTSDD1919 pKa = 3.11GSFIWKK1925 pKa = 8.11YY1926 pKa = 9.48TISLTNLQNIIVDD1939 pKa = 4.42NIQVEE1944 pKa = 4.32DD1945 pKa = 4.27DD1946 pKa = 3.49LSKK1949 pKa = 11.24VFTHH1953 pKa = 6.69GEE1955 pKa = 4.13SFKK1958 pKa = 10.12VTNITASGNLKK1969 pKa = 10.95ANGLYY1974 pKa = 10.6DD1975 pKa = 3.86GLNSMSTLLSGSYY1988 pKa = 9.8LAPLSKK1994 pKa = 10.69DD1995 pKa = 3.45SIVIEE2000 pKa = 4.2VQVYY2004 pKa = 7.27SHH2006 pKa = 6.37NFIGNVYY2013 pKa = 9.68NQALFEE2019 pKa = 4.43GFSNSTGQIKK2029 pKa = 10.1DD2030 pKa = 3.61AQSDD2034 pKa = 3.95DD2035 pKa = 3.99PLNTEE2040 pKa = 5.1ASPSSPRR2047 pKa = 11.84TTITFIPEE2055 pKa = 4.01VEE2057 pKa = 3.97LHH2059 pKa = 5.88IPEE2062 pKa = 4.68GFSPNKK2068 pKa = 10.35DD2069 pKa = 3.16MYY2071 pKa = 11.03NDD2073 pKa = 3.39TFEE2076 pKa = 4.57IVHH2079 pKa = 6.3SGNVTLEE2086 pKa = 3.9VEE2088 pKa = 4.55VFNRR2092 pKa = 11.84WGNRR2096 pKa = 11.84VYY2098 pKa = 10.93RR2099 pKa = 11.84STDD2102 pKa = 3.18YY2103 pKa = 11.42HH2104 pKa = 7.34DD2105 pKa = 5.03DD2106 pKa = 3.56WDD2108 pKa = 4.78GKK2110 pKa = 11.06GADD2113 pKa = 3.75NLLGKK2118 pKa = 9.77DD2119 pKa = 4.06LPEE2122 pKa = 3.81GTYY2125 pKa = 10.11FYY2127 pKa = 10.73IVTTTNKK2134 pKa = 8.57KK2135 pKa = 7.56TSEE2138 pKa = 3.98VKK2140 pKa = 10.3KK2141 pKa = 10.44IKK2143 pKa = 10.64GSVTLRR2149 pKa = 11.84RR2150 pKa = 3.8
Molecular weight: 221.38 kDa
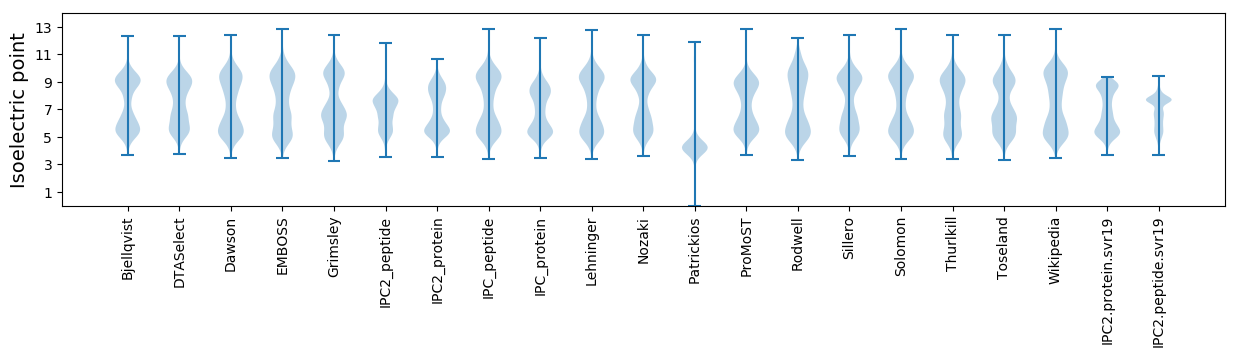
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A170YMZ3|A0A170YMZ3_9BACT Kef-type K+ transport system OS=Paludibacter jiangxiensis OX=681398 GN=PJIAN_1511 PE=4 SV=1
MM1 pKa = 7.64HH2 pKa = 7.33FPFDD6 pKa = 3.5VTINFAQSRR15 pKa = 11.84IPNVQNRR22 pKa = 11.84TIKK25 pKa = 10.27VYY27 pKa = 11.1NGITTFLGPNGSGKK41 pKa = 7.79TQLLRR46 pKa = 11.84GLKK49 pKa = 9.13TSLNIHH55 pKa = 6.5LNGKK59 pKa = 8.72KK60 pKa = 9.4IRR62 pKa = 11.84YY63 pKa = 8.64ISAGRR68 pKa = 11.84LGVMEE73 pKa = 5.39RR74 pKa = 11.84YY75 pKa = 9.92RR76 pKa = 11.84SDD78 pKa = 3.2FDD80 pKa = 2.91GWRR83 pKa = 11.84NQTIDD88 pKa = 3.5FDD90 pKa = 4.16SAEE93 pKa = 4.35FGSKK97 pKa = 9.84GAVARR102 pKa = 11.84RR103 pKa = 11.84HH104 pKa = 6.37LLFQVCLMFSPCWRR118 pKa = 11.84RR119 pKa = 11.84FSDD122 pKa = 4.73SVTCAHH128 pKa = 6.1NQRR131 pKa = 11.84GLL133 pKa = 3.4
MM1 pKa = 7.64HH2 pKa = 7.33FPFDD6 pKa = 3.5VTINFAQSRR15 pKa = 11.84IPNVQNRR22 pKa = 11.84TIKK25 pKa = 10.27VYY27 pKa = 11.1NGITTFLGPNGSGKK41 pKa = 7.79TQLLRR46 pKa = 11.84GLKK49 pKa = 9.13TSLNIHH55 pKa = 6.5LNGKK59 pKa = 8.72KK60 pKa = 9.4IRR62 pKa = 11.84YY63 pKa = 8.64ISAGRR68 pKa = 11.84LGVMEE73 pKa = 5.39RR74 pKa = 11.84YY75 pKa = 9.92RR76 pKa = 11.84SDD78 pKa = 3.2FDD80 pKa = 2.91GWRR83 pKa = 11.84NQTIDD88 pKa = 3.5FDD90 pKa = 4.16SAEE93 pKa = 4.35FGSKK97 pKa = 9.84GAVARR102 pKa = 11.84RR103 pKa = 11.84HH104 pKa = 6.37LLFQVCLMFSPCWRR118 pKa = 11.84RR119 pKa = 11.84FSDD122 pKa = 4.73SVTCAHH128 pKa = 6.1NQRR131 pKa = 11.84GLL133 pKa = 3.4
Molecular weight: 15.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1080957 |
26 |
6762 |
367.0 |
41.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.426 ± 0.045 | 1.116 ± 0.017 |
5.266 ± 0.032 | 5.575 ± 0.058 |
4.787 ± 0.037 | 6.894 ± 0.05 |
1.855 ± 0.02 | 7.014 ± 0.045 |
6.809 ± 0.053 | 9.014 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.468 ± 0.024 | 5.461 ± 0.04 |
3.892 ± 0.023 | 3.589 ± 0.03 |
3.963 ± 0.034 | 6.715 ± 0.058 |
6.02 ± 0.088 | 6.658 ± 0.043 |
1.283 ± 0.021 | 4.195 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
