
Alfalfa associated nucleorhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus; unclassified Nucleorhabdovirus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
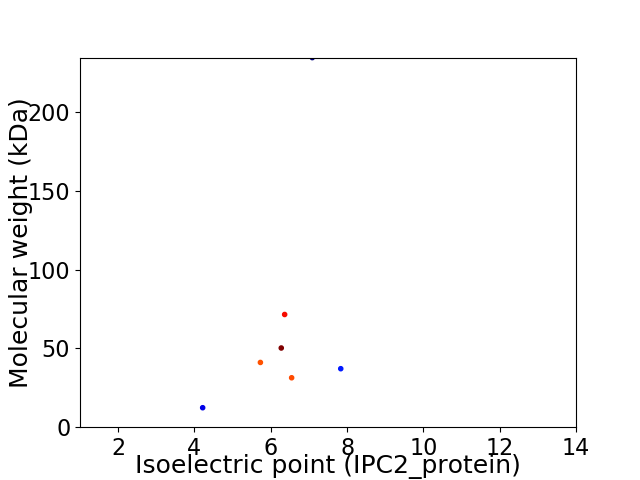
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
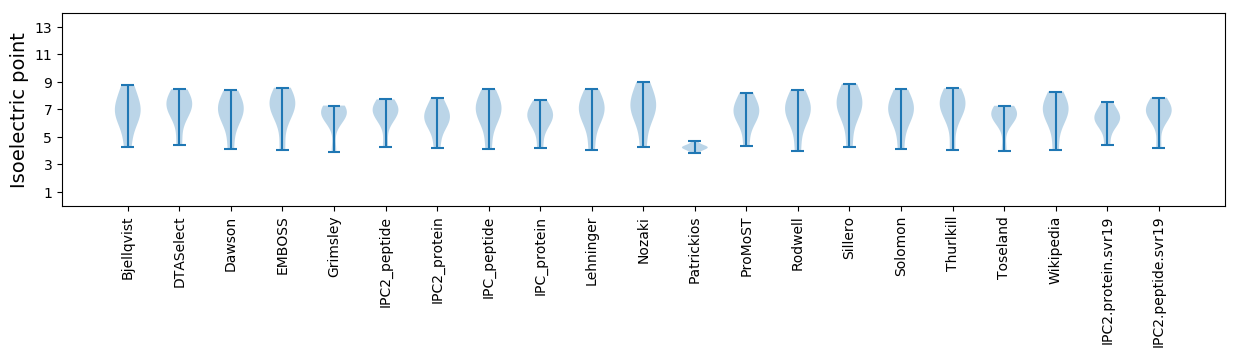
* You can choose from 21 different methods for calculating isoelectric point
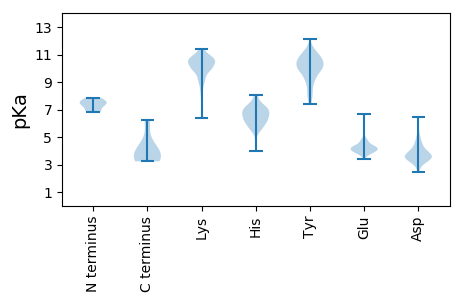
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A451G5G6|A0A451G5G6_9RHAB Phosphoprotein OS=Alfalfa associated nucleorhabdovirus OX=2518374 GN=P PE=4 SV=1
MM1 pKa = 6.8STTRR5 pKa = 11.84IDD7 pKa = 3.39NSGWDD12 pKa = 3.98SEE14 pKa = 4.7DD15 pKa = 3.56SMEE18 pKa = 4.45GVCFDD23 pKa = 3.52IHH25 pKa = 8.01DD26 pKa = 4.23GDD28 pKa = 4.16MIIPVKK34 pKa = 10.44FIINDD39 pKa = 3.88LVRR42 pKa = 11.84DD43 pKa = 3.81IFKK46 pKa = 11.07APDD49 pKa = 3.24NSDD52 pKa = 4.02SIRR55 pKa = 11.84EE56 pKa = 3.88PTLNNKK62 pKa = 8.99AAEE65 pKa = 4.3HH66 pKa = 6.55GEE68 pKa = 4.01DD69 pKa = 3.82PEE71 pKa = 4.44KK72 pKa = 11.12KK73 pKa = 8.07EE74 pKa = 4.38TEE76 pKa = 4.27PTTHH80 pKa = 7.3DD81 pKa = 3.64SVHH84 pKa = 6.38EE85 pKa = 4.19KK86 pKa = 9.4MQTTEE91 pKa = 4.47PEE93 pKa = 4.08DD94 pKa = 3.68AAPTSPSVPCSGTEE108 pKa = 4.04SASSNN113 pKa = 3.38
MM1 pKa = 6.8STTRR5 pKa = 11.84IDD7 pKa = 3.39NSGWDD12 pKa = 3.98SEE14 pKa = 4.7DD15 pKa = 3.56SMEE18 pKa = 4.45GVCFDD23 pKa = 3.52IHH25 pKa = 8.01DD26 pKa = 4.23GDD28 pKa = 4.16MIIPVKK34 pKa = 10.44FIINDD39 pKa = 3.88LVRR42 pKa = 11.84DD43 pKa = 3.81IFKK46 pKa = 11.07APDD49 pKa = 3.24NSDD52 pKa = 4.02SIRR55 pKa = 11.84EE56 pKa = 3.88PTLNNKK62 pKa = 8.99AAEE65 pKa = 4.3HH66 pKa = 6.55GEE68 pKa = 4.01DD69 pKa = 3.82PEE71 pKa = 4.44KK72 pKa = 11.12KK73 pKa = 8.07EE74 pKa = 4.38TEE76 pKa = 4.27PTTHH80 pKa = 7.3DD81 pKa = 3.64SVHH84 pKa = 6.38EE85 pKa = 4.19KK86 pKa = 9.4MQTTEE91 pKa = 4.47PEE93 pKa = 4.08DD94 pKa = 3.68AAPTSPSVPCSGTEE108 pKa = 4.04SASSNN113 pKa = 3.38
Molecular weight: 12.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A410HY56|A0A410HY56_9RHAB Glycoprotein OS=Alfalfa associated nucleorhabdovirus OX=2518374 GN=G PE=4 SV=1
MM1 pKa = 6.9TAPRR5 pKa = 11.84TMNDD9 pKa = 2.98KK10 pKa = 10.97KK11 pKa = 10.95KK12 pKa = 10.16DD13 pKa = 3.5TLLPTKK19 pKa = 10.35KK20 pKa = 9.17RR21 pKa = 11.84TSQDD25 pKa = 2.55KK26 pKa = 9.76YY27 pKa = 10.28NFRR30 pKa = 11.84STEE33 pKa = 3.78SLYY36 pKa = 11.05AEE38 pKa = 5.28PYY40 pKa = 10.08NKK42 pKa = 9.69IIRR45 pKa = 11.84TKK47 pKa = 10.89DD48 pKa = 3.04CLLPQRR54 pKa = 11.84TLMWFLSLTDD64 pKa = 3.32QSIEE68 pKa = 3.88TRR70 pKa = 11.84IEE72 pKa = 3.95KK73 pKa = 10.26IKK75 pKa = 9.57ITWHH79 pKa = 6.37PSVLPDD85 pKa = 3.76PKK87 pKa = 10.72KK88 pKa = 10.48KK89 pKa = 9.26IHH91 pKa = 6.9IMIKK95 pKa = 10.16DD96 pKa = 3.44RR97 pKa = 11.84EE98 pKa = 4.33EE99 pKa = 4.41YY100 pKa = 10.14SVHH103 pKa = 6.89SSAIYY108 pKa = 10.48SNGDD112 pKa = 3.1NTVLHH117 pKa = 6.58AVGLLSEE124 pKa = 4.45MVCVEE129 pKa = 4.09WSPCHH134 pKa = 7.15SYY136 pKa = 10.5ILEE139 pKa = 4.44RR140 pKa = 11.84NDD142 pKa = 3.25RR143 pKa = 11.84TILPWVVRR151 pKa = 11.84IEE153 pKa = 3.83HH154 pKa = 6.27DD155 pKa = 3.39QRR157 pKa = 11.84SVLEE161 pKa = 4.71DD162 pKa = 3.39GVSFGTVEE170 pKa = 3.67ITGVIHH176 pKa = 7.0CARR179 pKa = 11.84SHH181 pKa = 6.12KK182 pKa = 10.22GVPRR186 pKa = 11.84TIPFSEE192 pKa = 3.95PRR194 pKa = 11.84VLGSNINFPYY204 pKa = 10.38YY205 pKa = 10.19SPCFIGRR212 pKa = 11.84GARR215 pKa = 11.84SNYY218 pKa = 9.22KK219 pKa = 10.29RR220 pKa = 11.84IGVSNCQEE228 pKa = 3.75DD229 pKa = 4.1AFEE232 pKa = 4.97KK233 pKa = 10.61EE234 pKa = 3.81IMPYY238 pKa = 9.92LPKK241 pKa = 10.6DD242 pKa = 3.49ISPKK246 pKa = 10.58GLWKK250 pKa = 10.37LKK252 pKa = 10.67CLITDD257 pKa = 3.08QDD259 pKa = 3.3KK260 pKa = 11.26RR261 pKa = 11.84DD262 pKa = 3.98IISCITSCRR271 pKa = 11.84YY272 pKa = 9.99DD273 pKa = 3.51RR274 pKa = 11.84GTMNCNCDD282 pKa = 3.15RR283 pKa = 11.84VISTKK288 pKa = 10.64LLAIRR293 pKa = 11.84GSFLTPYY300 pKa = 10.09IEE302 pKa = 4.25WDD304 pKa = 3.67QVVHH308 pKa = 7.82DD309 pKa = 4.53ITTDD313 pKa = 3.11KK314 pKa = 10.87QSPYY318 pKa = 11.01DD319 pKa = 3.73LPTT322 pKa = 3.53
MM1 pKa = 6.9TAPRR5 pKa = 11.84TMNDD9 pKa = 2.98KK10 pKa = 10.97KK11 pKa = 10.95KK12 pKa = 10.16DD13 pKa = 3.5TLLPTKK19 pKa = 10.35KK20 pKa = 9.17RR21 pKa = 11.84TSQDD25 pKa = 2.55KK26 pKa = 9.76YY27 pKa = 10.28NFRR30 pKa = 11.84STEE33 pKa = 3.78SLYY36 pKa = 11.05AEE38 pKa = 5.28PYY40 pKa = 10.08NKK42 pKa = 9.69IIRR45 pKa = 11.84TKK47 pKa = 10.89DD48 pKa = 3.04CLLPQRR54 pKa = 11.84TLMWFLSLTDD64 pKa = 3.32QSIEE68 pKa = 3.88TRR70 pKa = 11.84IEE72 pKa = 3.95KK73 pKa = 10.26IKK75 pKa = 9.57ITWHH79 pKa = 6.37PSVLPDD85 pKa = 3.76PKK87 pKa = 10.72KK88 pKa = 10.48KK89 pKa = 9.26IHH91 pKa = 6.9IMIKK95 pKa = 10.16DD96 pKa = 3.44RR97 pKa = 11.84EE98 pKa = 4.33EE99 pKa = 4.41YY100 pKa = 10.14SVHH103 pKa = 6.89SSAIYY108 pKa = 10.48SNGDD112 pKa = 3.1NTVLHH117 pKa = 6.58AVGLLSEE124 pKa = 4.45MVCVEE129 pKa = 4.09WSPCHH134 pKa = 7.15SYY136 pKa = 10.5ILEE139 pKa = 4.44RR140 pKa = 11.84NDD142 pKa = 3.25RR143 pKa = 11.84TILPWVVRR151 pKa = 11.84IEE153 pKa = 3.83HH154 pKa = 6.27DD155 pKa = 3.39QRR157 pKa = 11.84SVLEE161 pKa = 4.71DD162 pKa = 3.39GVSFGTVEE170 pKa = 3.67ITGVIHH176 pKa = 7.0CARR179 pKa = 11.84SHH181 pKa = 6.12KK182 pKa = 10.22GVPRR186 pKa = 11.84TIPFSEE192 pKa = 3.95PRR194 pKa = 11.84VLGSNINFPYY204 pKa = 10.38YY205 pKa = 10.19SPCFIGRR212 pKa = 11.84GARR215 pKa = 11.84SNYY218 pKa = 9.22KK219 pKa = 10.29RR220 pKa = 11.84IGVSNCQEE228 pKa = 3.75DD229 pKa = 4.1AFEE232 pKa = 4.97KK233 pKa = 10.61EE234 pKa = 3.81IMPYY238 pKa = 9.92LPKK241 pKa = 10.6DD242 pKa = 3.49ISPKK246 pKa = 10.58GLWKK250 pKa = 10.37LKK252 pKa = 10.67CLITDD257 pKa = 3.08QDD259 pKa = 3.3KK260 pKa = 11.26RR261 pKa = 11.84DD262 pKa = 3.98IISCITSCRR271 pKa = 11.84YY272 pKa = 9.99DD273 pKa = 3.51RR274 pKa = 11.84GTMNCNCDD282 pKa = 3.15RR283 pKa = 11.84VISTKK288 pKa = 10.64LLAIRR293 pKa = 11.84GSFLTPYY300 pKa = 10.09IEE302 pKa = 4.25WDD304 pKa = 3.67QVVHH308 pKa = 7.82DD309 pKa = 4.53ITTDD313 pKa = 3.11KK314 pKa = 10.87QSPYY318 pKa = 11.01DD319 pKa = 3.73LPTT322 pKa = 3.53
Molecular weight: 37.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4185 |
113 |
2038 |
597.9 |
68.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.349 ± 0.48 | 1.983 ± 0.286 |
6.523 ± 0.296 | 6.786 ± 0.438 |
3.297 ± 0.568 | 4.851 ± 0.121 |
2.7 ± 0.17 | 8.363 ± 0.29 |
8.005 ± 0.213 | 8.196 ± 1.082 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.465 ± 0.2 | 4.683 ± 0.313 |
3.56 ± 0.482 | 2.413 ± 0.346 |
5.018 ± 0.26 | 8.984 ± 0.355 |
5.926 ± 0.527 | 5.52 ± 0.685 |
1.338 ± 0.188 | 4.038 ± 0.421 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
