
Fusarium culmorum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales;
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
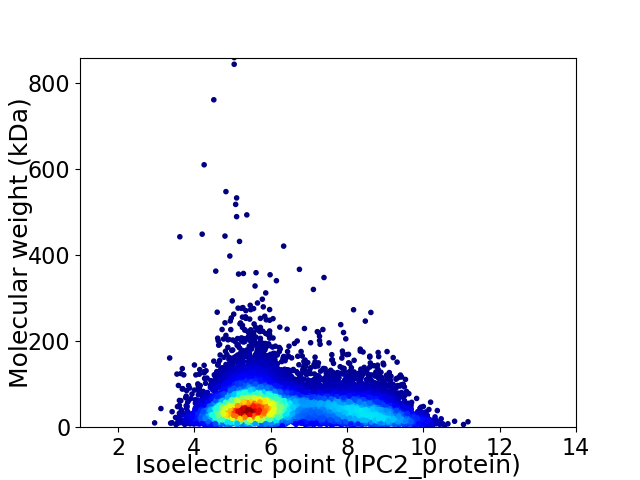
Virtual 2D-PAGE plot for 12360 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T4GEL4|A0A2T4GEL4_FUSCU Uncharacterized protein OS=Fusarium culmorum OX=5516 GN=FCULG_00010253 PE=4 SV=1
MM1 pKa = 7.82AYY3 pKa = 10.66SLTTSFIGEE12 pKa = 4.16SLLSGFEE19 pKa = 4.16WFNGTDD25 pKa = 3.4LSKK28 pKa = 11.38GFVQYY33 pKa = 11.13QDD35 pKa = 3.2FTGAFEE41 pKa = 4.39NGLYY45 pKa = 10.65SVDD48 pKa = 3.8PFSEE52 pKa = 4.77TVQLRR57 pKa = 11.84PDD59 pKa = 3.29SRR61 pKa = 11.84HH62 pKa = 6.33KK63 pKa = 11.06YY64 pKa = 10.08GLDD67 pKa = 3.0EE68 pKa = 4.46GRR70 pKa = 11.84PSIRR74 pKa = 11.84LEE76 pKa = 4.12SKK78 pKa = 10.14EE79 pKa = 4.04SYY81 pKa = 10.47QYY83 pKa = 11.98GLFIADD89 pKa = 4.16FQHH92 pKa = 6.63MPISQCGTWPAFWAYY107 pKa = 9.91GSNWPMGGEE116 pKa = 3.82IDD118 pKa = 3.63ILEE121 pKa = 4.64GANLAYY127 pKa = 9.51TNIMSAHH134 pKa = 5.16TAEE137 pKa = 4.57GCMLDD142 pKa = 3.98PADD145 pKa = 4.36SNLFSGIRR153 pKa = 11.84RR154 pKa = 11.84NLDD157 pKa = 3.17CGVGTNNVGCGFNPPKK173 pKa = 10.45SDD175 pKa = 2.97TSSYY179 pKa = 11.36GDD181 pKa = 3.35GFNAVGGGVYY191 pKa = 10.63AMEE194 pKa = 4.64WDD196 pKa = 3.6SEE198 pKa = 4.75YY199 pKa = 10.73ISIWHH204 pKa = 6.56FPRR207 pKa = 11.84GAIPADD213 pKa = 3.51IEE215 pKa = 4.53AKK217 pKa = 10.53QPDD220 pKa = 4.02PRR222 pKa = 11.84NWGLPQSLFGGAKK235 pKa = 9.89CDD237 pKa = 3.27VDD239 pKa = 5.41KK240 pKa = 11.48YY241 pKa = 10.76FSDD244 pKa = 3.35MKK246 pKa = 10.93LVLNINFCGDD256 pKa = 3.43YY257 pKa = 11.63GEE259 pKa = 4.7GTWANSKK266 pKa = 9.28VCRR269 pKa = 11.84ALAPTCRR276 pKa = 11.84EE277 pKa = 3.86YY278 pKa = 11.3VANNPTAFKK287 pKa = 10.12EE288 pKa = 4.27AYY290 pKa = 9.86FDD292 pKa = 3.29VSYY295 pKa = 11.1IDD297 pKa = 3.24VYY299 pKa = 11.02TRR301 pKa = 11.84LGGNIPPVLPSSASEE316 pKa = 4.05ASTQASDD323 pKa = 3.69VIIPGEE329 pKa = 4.22SATNTPMSGRR339 pKa = 11.84PVFPSTNFPGNSTIATRR356 pKa = 11.84TDD358 pKa = 3.09RR359 pKa = 11.84KK360 pKa = 10.75GKK362 pKa = 8.6ATQSEE367 pKa = 5.02EE368 pKa = 4.1PTTTTTLTMTGISSVLVTIPGSGTDD393 pKa = 3.71TPTVTPLPVATGGKK407 pKa = 9.0SANPSKK413 pKa = 10.74IGDD416 pKa = 3.7YY417 pKa = 10.88SYY419 pKa = 11.3LGCFGSQNGFQTFDD433 pKa = 3.88LADD436 pKa = 4.78DD437 pKa = 4.29SDD439 pKa = 4.34DD440 pKa = 3.69MTIEE444 pKa = 4.12RR445 pKa = 11.84CVDD448 pKa = 2.91ACNGLTYY455 pKa = 10.29IGLFEE460 pKa = 4.26GTCYY464 pKa = 10.14CASVLDD470 pKa = 4.31GDD472 pKa = 4.1TLAIRR477 pKa = 11.84NEE479 pKa = 4.32TSCNRR484 pKa = 11.84PCPGDD489 pKa = 3.77DD490 pKa = 3.42AQFCGGMVTQNTKK503 pKa = 8.16TRR505 pKa = 11.84FRR507 pKa = 11.84RR508 pKa = 11.84TTPLRR513 pKa = 11.84RR514 pKa = 11.84DD515 pKa = 3.31APSNVLLTVYY525 pKa = 11.16ADD527 pKa = 3.32ISDD530 pKa = 4.02VEE532 pKa = 4.38LPEE535 pKa = 4.35VPPAMGPGTNDD546 pKa = 2.69TSTDD550 pKa = 3.67DD551 pKa = 4.41GDD553 pKa = 4.18SQGDD557 pKa = 3.88SATDD561 pKa = 3.42EE562 pKa = 4.63SEE564 pKa = 4.58SPTTSDD570 pKa = 4.69DD571 pKa = 3.61NASQGASDD579 pKa = 3.51AAEE582 pKa = 4.54PEE584 pKa = 4.01NSGDD588 pKa = 3.69SDD590 pKa = 3.8VTTSQEE596 pKa = 3.81STNADD601 pKa = 2.97GSEE604 pKa = 4.48DD605 pKa = 3.37LTASNDD611 pKa = 3.08NGTQGGSTTDD621 pKa = 4.01PDD623 pKa = 4.82DD624 pKa = 3.8STASDD629 pKa = 3.67GNASQRR635 pKa = 11.84GSEE638 pKa = 4.19SGDD641 pKa = 3.52SKK643 pKa = 10.94ILPVSNDD650 pKa = 3.53NIDD653 pKa = 3.4ASLAGSASSPGSSAQGSSDD672 pKa = 3.69EE673 pKa = 4.51QDD675 pKa = 3.4SSDD678 pKa = 4.94DD679 pKa = 3.85QDD681 pKa = 5.9DD682 pKa = 4.13PDD684 pKa = 4.87SDD686 pKa = 4.22GNSILPSAAGVVLSDD701 pKa = 4.91DD702 pKa = 3.47QDD704 pKa = 3.95TPPVAVIGTASDD716 pKa = 3.86GLVLGATQSAADD728 pKa = 3.84SNQEE732 pKa = 3.8LGIITMSYY740 pKa = 9.57EE741 pKa = 3.67KK742 pKa = 10.21CDD744 pKa = 3.57YY745 pKa = 11.21CEE747 pKa = 3.88TPEE750 pKa = 4.83LIKK753 pKa = 10.74PPMEE757 pKa = 4.28TKK759 pKa = 10.58VVDD762 pKa = 4.44CDD764 pKa = 3.59GCGPNGEE771 pKa = 4.36DD772 pKa = 3.5TVTLTVPISVTVTVPGASTVQAQQTEE798 pKa = 4.27KK799 pKa = 10.76AAAEE803 pKa = 4.12AVTSYY808 pKa = 11.18GDD810 pKa = 3.82SPEE813 pKa = 3.95QPPRR817 pKa = 11.84NSTILPVVPNGMGSGAEE834 pKa = 3.91AGEE837 pKa = 4.41SPEE840 pKa = 3.92VTTVVITYY848 pKa = 7.5LTTQLVTYY856 pKa = 7.47GTSTDD861 pKa = 3.39SVSTHH866 pKa = 6.3LAYY869 pKa = 9.67PIPGQPNTPVTIATPTPGAPSEE891 pKa = 4.1PVAVSAASSRR901 pKa = 11.84RR902 pKa = 11.84DD903 pKa = 3.36DD904 pKa = 3.56VFVYY908 pKa = 8.89FAIVAMAILALTLL921 pKa = 3.7
MM1 pKa = 7.82AYY3 pKa = 10.66SLTTSFIGEE12 pKa = 4.16SLLSGFEE19 pKa = 4.16WFNGTDD25 pKa = 3.4LSKK28 pKa = 11.38GFVQYY33 pKa = 11.13QDD35 pKa = 3.2FTGAFEE41 pKa = 4.39NGLYY45 pKa = 10.65SVDD48 pKa = 3.8PFSEE52 pKa = 4.77TVQLRR57 pKa = 11.84PDD59 pKa = 3.29SRR61 pKa = 11.84HH62 pKa = 6.33KK63 pKa = 11.06YY64 pKa = 10.08GLDD67 pKa = 3.0EE68 pKa = 4.46GRR70 pKa = 11.84PSIRR74 pKa = 11.84LEE76 pKa = 4.12SKK78 pKa = 10.14EE79 pKa = 4.04SYY81 pKa = 10.47QYY83 pKa = 11.98GLFIADD89 pKa = 4.16FQHH92 pKa = 6.63MPISQCGTWPAFWAYY107 pKa = 9.91GSNWPMGGEE116 pKa = 3.82IDD118 pKa = 3.63ILEE121 pKa = 4.64GANLAYY127 pKa = 9.51TNIMSAHH134 pKa = 5.16TAEE137 pKa = 4.57GCMLDD142 pKa = 3.98PADD145 pKa = 4.36SNLFSGIRR153 pKa = 11.84RR154 pKa = 11.84NLDD157 pKa = 3.17CGVGTNNVGCGFNPPKK173 pKa = 10.45SDD175 pKa = 2.97TSSYY179 pKa = 11.36GDD181 pKa = 3.35GFNAVGGGVYY191 pKa = 10.63AMEE194 pKa = 4.64WDD196 pKa = 3.6SEE198 pKa = 4.75YY199 pKa = 10.73ISIWHH204 pKa = 6.56FPRR207 pKa = 11.84GAIPADD213 pKa = 3.51IEE215 pKa = 4.53AKK217 pKa = 10.53QPDD220 pKa = 4.02PRR222 pKa = 11.84NWGLPQSLFGGAKK235 pKa = 9.89CDD237 pKa = 3.27VDD239 pKa = 5.41KK240 pKa = 11.48YY241 pKa = 10.76FSDD244 pKa = 3.35MKK246 pKa = 10.93LVLNINFCGDD256 pKa = 3.43YY257 pKa = 11.63GEE259 pKa = 4.7GTWANSKK266 pKa = 9.28VCRR269 pKa = 11.84ALAPTCRR276 pKa = 11.84EE277 pKa = 3.86YY278 pKa = 11.3VANNPTAFKK287 pKa = 10.12EE288 pKa = 4.27AYY290 pKa = 9.86FDD292 pKa = 3.29VSYY295 pKa = 11.1IDD297 pKa = 3.24VYY299 pKa = 11.02TRR301 pKa = 11.84LGGNIPPVLPSSASEE316 pKa = 4.05ASTQASDD323 pKa = 3.69VIIPGEE329 pKa = 4.22SATNTPMSGRR339 pKa = 11.84PVFPSTNFPGNSTIATRR356 pKa = 11.84TDD358 pKa = 3.09RR359 pKa = 11.84KK360 pKa = 10.75GKK362 pKa = 8.6ATQSEE367 pKa = 5.02EE368 pKa = 4.1PTTTTTLTMTGISSVLVTIPGSGTDD393 pKa = 3.71TPTVTPLPVATGGKK407 pKa = 9.0SANPSKK413 pKa = 10.74IGDD416 pKa = 3.7YY417 pKa = 10.88SYY419 pKa = 11.3LGCFGSQNGFQTFDD433 pKa = 3.88LADD436 pKa = 4.78DD437 pKa = 4.29SDD439 pKa = 4.34DD440 pKa = 3.69MTIEE444 pKa = 4.12RR445 pKa = 11.84CVDD448 pKa = 2.91ACNGLTYY455 pKa = 10.29IGLFEE460 pKa = 4.26GTCYY464 pKa = 10.14CASVLDD470 pKa = 4.31GDD472 pKa = 4.1TLAIRR477 pKa = 11.84NEE479 pKa = 4.32TSCNRR484 pKa = 11.84PCPGDD489 pKa = 3.77DD490 pKa = 3.42AQFCGGMVTQNTKK503 pKa = 8.16TRR505 pKa = 11.84FRR507 pKa = 11.84RR508 pKa = 11.84TTPLRR513 pKa = 11.84RR514 pKa = 11.84DD515 pKa = 3.31APSNVLLTVYY525 pKa = 11.16ADD527 pKa = 3.32ISDD530 pKa = 4.02VEE532 pKa = 4.38LPEE535 pKa = 4.35VPPAMGPGTNDD546 pKa = 2.69TSTDD550 pKa = 3.67DD551 pKa = 4.41GDD553 pKa = 4.18SQGDD557 pKa = 3.88SATDD561 pKa = 3.42EE562 pKa = 4.63SEE564 pKa = 4.58SPTTSDD570 pKa = 4.69DD571 pKa = 3.61NASQGASDD579 pKa = 3.51AAEE582 pKa = 4.54PEE584 pKa = 4.01NSGDD588 pKa = 3.69SDD590 pKa = 3.8VTTSQEE596 pKa = 3.81STNADD601 pKa = 2.97GSEE604 pKa = 4.48DD605 pKa = 3.37LTASNDD611 pKa = 3.08NGTQGGSTTDD621 pKa = 4.01PDD623 pKa = 4.82DD624 pKa = 3.8STASDD629 pKa = 3.67GNASQRR635 pKa = 11.84GSEE638 pKa = 4.19SGDD641 pKa = 3.52SKK643 pKa = 10.94ILPVSNDD650 pKa = 3.53NIDD653 pKa = 3.4ASLAGSASSPGSSAQGSSDD672 pKa = 3.69EE673 pKa = 4.51QDD675 pKa = 3.4SSDD678 pKa = 4.94DD679 pKa = 3.85QDD681 pKa = 5.9DD682 pKa = 4.13PDD684 pKa = 4.87SDD686 pKa = 4.22GNSILPSAAGVVLSDD701 pKa = 4.91DD702 pKa = 3.47QDD704 pKa = 3.95TPPVAVIGTASDD716 pKa = 3.86GLVLGATQSAADD728 pKa = 3.84SNQEE732 pKa = 3.8LGIITMSYY740 pKa = 9.57EE741 pKa = 3.67KK742 pKa = 10.21CDD744 pKa = 3.57YY745 pKa = 11.21CEE747 pKa = 3.88TPEE750 pKa = 4.83LIKK753 pKa = 10.74PPMEE757 pKa = 4.28TKK759 pKa = 10.58VVDD762 pKa = 4.44CDD764 pKa = 3.59GCGPNGEE771 pKa = 4.36DD772 pKa = 3.5TVTLTVPISVTVTVPGASTVQAQQTEE798 pKa = 4.27KK799 pKa = 10.76AAAEE803 pKa = 4.12AVTSYY808 pKa = 11.18GDD810 pKa = 3.82SPEE813 pKa = 3.95QPPRR817 pKa = 11.84NSTILPVVPNGMGSGAEE834 pKa = 3.91AGEE837 pKa = 4.41SPEE840 pKa = 3.92VTTVVITYY848 pKa = 7.5LTTQLVTYY856 pKa = 7.47GTSTDD861 pKa = 3.39SVSTHH866 pKa = 6.3LAYY869 pKa = 9.67PIPGQPNTPVTIATPTPGAPSEE891 pKa = 4.1PVAVSAASSRR901 pKa = 11.84RR902 pKa = 11.84DD903 pKa = 3.36DD904 pKa = 3.56VFVYY908 pKa = 8.89FAIVAMAILALTLL921 pKa = 3.7
Molecular weight: 96.56 kDa
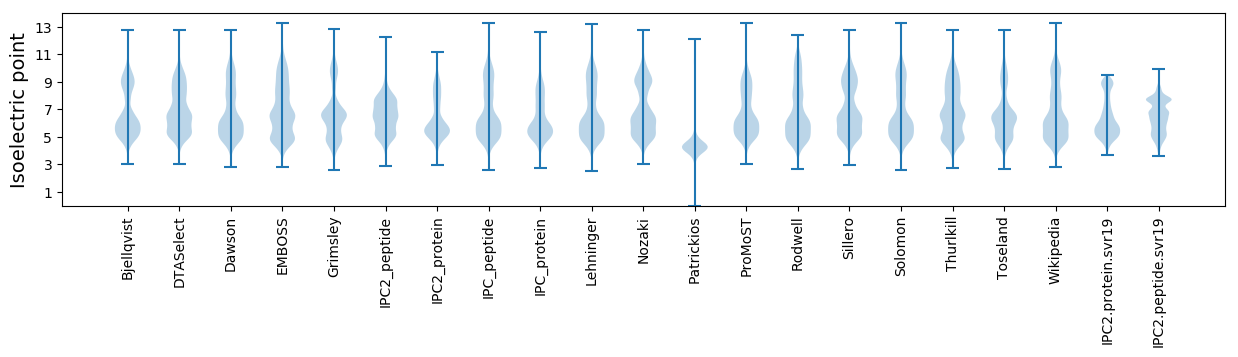
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T4GI23|A0A2T4GI23_FUSCU ATPase GET3 OS=Fusarium culmorum OX=5516 GN=GET3 PE=3 SV=1
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.63RR8 pKa = 11.84HH9 pKa = 4.04TAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APARR26 pKa = 11.84SNRR29 pKa = 11.84HH30 pKa = 4.32TVTTTTTTTTKK41 pKa = 9.91PRR43 pKa = 11.84RR44 pKa = 11.84GMFGGGSTARR54 pKa = 11.84RR55 pKa = 11.84THH57 pKa = 5.96GAAPVHH63 pKa = 5.35HH64 pKa = 6.52HH65 pKa = 5.47QRR67 pKa = 11.84RR68 pKa = 11.84PSMKK72 pKa = 10.02DD73 pKa = 2.95KK74 pKa = 11.41VSGALLKK81 pKa = 11.04LKK83 pKa = 10.68GSLTRR88 pKa = 11.84RR89 pKa = 11.84PGVKK93 pKa = 9.89AAGTRR98 pKa = 11.84RR99 pKa = 11.84MHH101 pKa = 5.56GTNGRR106 pKa = 11.84GARR109 pKa = 11.84HH110 pKa = 5.8HH111 pKa = 7.22RR112 pKa = 11.84YY113 pKa = 9.44
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.63RR8 pKa = 11.84HH9 pKa = 4.04TAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APARR26 pKa = 11.84SNRR29 pKa = 11.84HH30 pKa = 4.32TVTTTTTTTTKK41 pKa = 9.91PRR43 pKa = 11.84RR44 pKa = 11.84GMFGGGSTARR54 pKa = 11.84RR55 pKa = 11.84THH57 pKa = 5.96GAAPVHH63 pKa = 5.35HH64 pKa = 6.52HH65 pKa = 5.47QRR67 pKa = 11.84RR68 pKa = 11.84PSMKK72 pKa = 10.02DD73 pKa = 2.95KK74 pKa = 11.41VSGALLKK81 pKa = 11.04LKK83 pKa = 10.68GSLTRR88 pKa = 11.84RR89 pKa = 11.84PGVKK93 pKa = 9.89AAGTRR98 pKa = 11.84RR99 pKa = 11.84MHH101 pKa = 5.56GTNGRR106 pKa = 11.84GARR109 pKa = 11.84HH110 pKa = 5.8HH111 pKa = 7.22RR112 pKa = 11.84YY113 pKa = 9.44
Molecular weight: 12.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5813542 |
24 |
7826 |
470.4 |
52.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.269 ± 0.021 | 1.3 ± 0.01 |
5.936 ± 0.017 | 6.207 ± 0.026 |
3.769 ± 0.014 | 6.736 ± 0.02 |
2.362 ± 0.01 | 5.102 ± 0.016 |
5.124 ± 0.02 | 8.679 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.278 ± 0.009 | 3.862 ± 0.01 |
5.856 ± 0.023 | 4.015 ± 0.017 |
5.793 ± 0.02 | 8.129 ± 0.024 |
6.124 ± 0.029 | 6.15 ± 0.018 |
1.514 ± 0.009 | 2.796 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
