
Phreatobacter sp. NMCR1094
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phreatobacteraceae; Phreatobacter; unclassified Phreatobacter
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
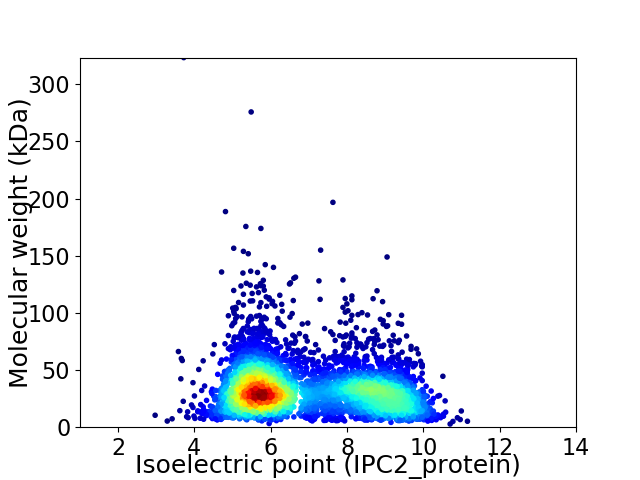
Virtual 2D-PAGE plot for 4643 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D7QJS5|A0A4D7QJS5_9PROT ABC transporter permease OS=Phreatobacter sp. NMCR1094 OX=2570229 GN=E8L99_07135 PE=3 SV=1
MM1 pKa = 7.29SLSGALNSAVSALSSQSSALAMISDD26 pKa = 5.01NISNSSTYY34 pKa = 10.57GYY36 pKa = 8.0KK37 pKa = 8.19TTTASFEE44 pKa = 4.42SMLTGSSSSAYY55 pKa = 9.8SSGGVSVSSVSNISTAGLLVASTTSSNMAISGSGFFVVTDD95 pKa = 4.38GANSNQTFYY104 pKa = 11.51SRR106 pKa = 11.84NGEE109 pKa = 4.04FTTDD113 pKa = 2.79SDD115 pKa = 4.54GYY117 pKa = 10.72LVNGDD122 pKa = 4.45YY123 pKa = 11.04YY124 pKa = 10.84LQGWKK129 pKa = 8.79TDD131 pKa = 3.33AEE133 pKa = 4.49GNIVGSSTQSSLTAIDD149 pKa = 3.9TNSVATIASATTKK162 pKa = 10.13MSLQANLPSNAATNDD177 pKa = 3.44TFTSEE182 pKa = 4.06VEE184 pKa = 4.49LYY186 pKa = 10.81DD187 pKa = 4.15SLGTAATSTVTWTKK201 pKa = 10.35TGDD204 pKa = 3.96NAWTASFSDD213 pKa = 3.64PTAADD218 pKa = 3.51GTTQLGTVSSSDD230 pKa = 2.91ITITFNTDD238 pKa = 2.01GTLASTSPSPATLSIGAWTTGAADD262 pKa = 3.51STITLDD268 pKa = 2.97MGTIGSSSGLTQLSSTADD286 pKa = 3.48TLAVSLTQDD295 pKa = 2.87QDD297 pKa = 3.55GVSYY301 pKa = 11.3GSLTGVEE308 pKa = 4.18IGDD311 pKa = 3.8NGTVYY316 pKa = 11.1ANYY319 pKa = 10.74DD320 pKa = 3.39NGEE323 pKa = 3.45QRR325 pKa = 11.84AIYY328 pKa = 9.02KK329 pKa = 9.85VAVATFNNADD339 pKa = 3.82GLTAMSGGVYY349 pKa = 9.95AASSEE354 pKa = 4.58SGGSTLHH361 pKa = 6.5IAGTGGAGDD370 pKa = 3.66VLGSKK375 pKa = 10.63LEE377 pKa = 4.17ASTTDD382 pKa = 3.27TNQEE386 pKa = 3.95FSNMMSAQQAYY397 pKa = 9.43SAAAQVMSTVNKK409 pKa = 10.0MYY411 pKa = 8.48DD412 pKa = 3.46TLISAVRR419 pKa = 3.41
MM1 pKa = 7.29SLSGALNSAVSALSSQSSALAMISDD26 pKa = 5.01NISNSSTYY34 pKa = 10.57GYY36 pKa = 8.0KK37 pKa = 8.19TTTASFEE44 pKa = 4.42SMLTGSSSSAYY55 pKa = 9.8SSGGVSVSSVSNISTAGLLVASTTSSNMAISGSGFFVVTDD95 pKa = 4.38GANSNQTFYY104 pKa = 11.51SRR106 pKa = 11.84NGEE109 pKa = 4.04FTTDD113 pKa = 2.79SDD115 pKa = 4.54GYY117 pKa = 10.72LVNGDD122 pKa = 4.45YY123 pKa = 11.04YY124 pKa = 10.84LQGWKK129 pKa = 8.79TDD131 pKa = 3.33AEE133 pKa = 4.49GNIVGSSTQSSLTAIDD149 pKa = 3.9TNSVATIASATTKK162 pKa = 10.13MSLQANLPSNAATNDD177 pKa = 3.44TFTSEE182 pKa = 4.06VEE184 pKa = 4.49LYY186 pKa = 10.81DD187 pKa = 4.15SLGTAATSTVTWTKK201 pKa = 10.35TGDD204 pKa = 3.96NAWTASFSDD213 pKa = 3.64PTAADD218 pKa = 3.51GTTQLGTVSSSDD230 pKa = 2.91ITITFNTDD238 pKa = 2.01GTLASTSPSPATLSIGAWTTGAADD262 pKa = 3.51STITLDD268 pKa = 2.97MGTIGSSSGLTQLSSTADD286 pKa = 3.48TLAVSLTQDD295 pKa = 2.87QDD297 pKa = 3.55GVSYY301 pKa = 11.3GSLTGVEE308 pKa = 4.18IGDD311 pKa = 3.8NGTVYY316 pKa = 11.1ANYY319 pKa = 10.74DD320 pKa = 3.39NGEE323 pKa = 3.45QRR325 pKa = 11.84AIYY328 pKa = 9.02KK329 pKa = 9.85VAVATFNNADD339 pKa = 3.82GLTAMSGGVYY349 pKa = 9.95AASSEE354 pKa = 4.58SGGSTLHH361 pKa = 6.5IAGTGGAGDD370 pKa = 3.66VLGSKK375 pKa = 10.63LEE377 pKa = 4.17ASTTDD382 pKa = 3.27TNQEE386 pKa = 3.95FSNMMSAQQAYY397 pKa = 9.43SAAAQVMSTVNKK409 pKa = 10.0MYY411 pKa = 8.48DD412 pKa = 3.46TLISAVRR419 pKa = 3.41
Molecular weight: 42.31 kDa
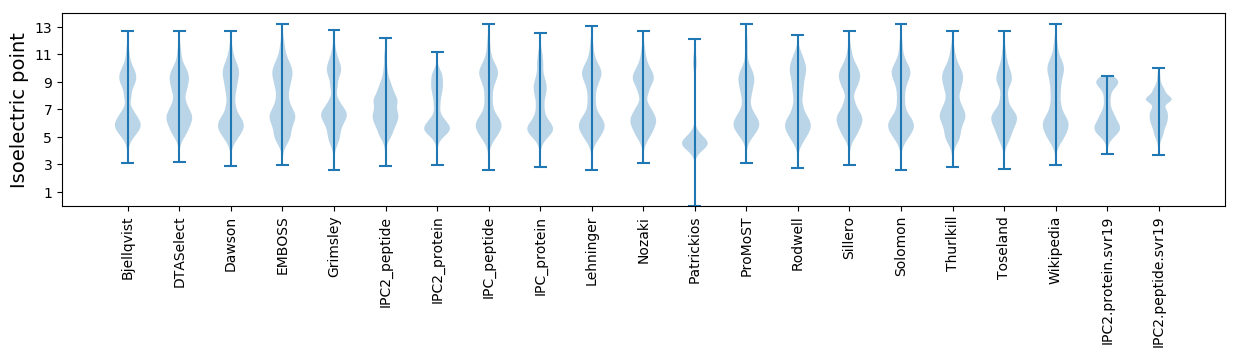
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D7QFR8|A0A4D7QFR8_9PROT Glutathione S-transferase OS=Phreatobacter sp. NMCR1094 OX=2570229 GN=E8L99_13900 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.69GGQRR29 pKa = 11.84ILSARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.69GGQRR29 pKa = 11.84ILSARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1481849 |
27 |
3288 |
319.2 |
34.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.256 ± 0.051 | 0.784 ± 0.011 |
5.426 ± 0.027 | 5.126 ± 0.028 |
3.73 ± 0.024 | 8.834 ± 0.031 |
1.928 ± 0.019 | 5.345 ± 0.026 |
2.99 ± 0.03 | 9.958 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.576 ± 0.018 | 2.376 ± 0.019 |
5.47 ± 0.028 | 3.036 ± 0.027 |
7.31 ± 0.036 | 5.239 ± 0.029 |
5.554 ± 0.041 | 7.748 ± 0.03 |
1.299 ± 0.012 | 2.014 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
