
Monoglobus pectinilyticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Monoglobus
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
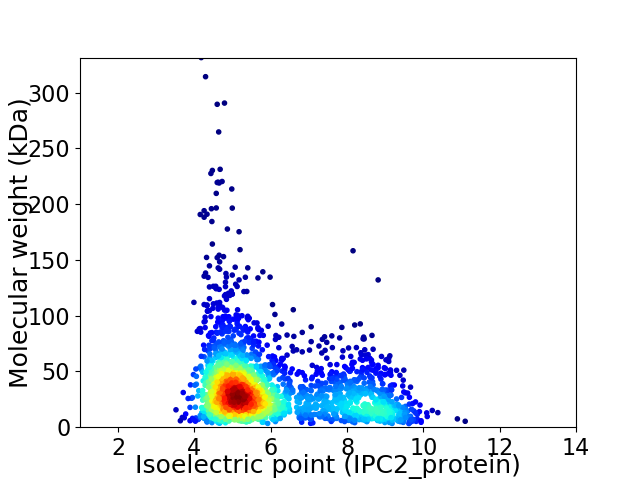
Virtual 2D-PAGE plot for 2258 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K9P291|A0A2K9P291_9FIRM GCN5-related N-acetyltransferase OS=Monoglobus pectinilyticus OX=1981510 GN=B9O19_01221 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84NLKK5 pKa = 10.19KK6 pKa = 10.71VIALIAVFAMMVSTVAFAATFGDD29 pKa = 3.96VKK31 pKa = 11.03EE32 pKa = 4.18GDD34 pKa = 3.58NYY36 pKa = 11.26YY37 pKa = 10.78EE38 pKa = 5.57AIEE41 pKa = 4.18TLNKK45 pKa = 10.47LGILTGDD52 pKa = 4.06DD53 pKa = 3.79EE54 pKa = 5.62NNDD57 pKa = 2.72GVMEE61 pKa = 3.84YY62 pKa = 10.36RR63 pKa = 11.84PNDD66 pKa = 3.91SITRR70 pKa = 11.84AEE72 pKa = 3.79ITVIVARR79 pKa = 11.84IQGQTGAVAQADD91 pKa = 4.1TVFTDD96 pKa = 3.55VPSTYY101 pKa = 9.23WASGYY106 pKa = 9.59IASATNQGIINGYY119 pKa = 10.16GDD121 pKa = 3.86GTFGPDD127 pKa = 3.96DD128 pKa = 3.64KK129 pKa = 11.59VLYY132 pKa = 10.29EE133 pKa = 4.67DD134 pKa = 4.02VIKK137 pKa = 10.51MLMEE141 pKa = 3.88TLGYY145 pKa = 10.01KK146 pKa = 10.19PYY148 pKa = 10.43AQNNGGYY155 pKa = 6.11PTGYY159 pKa = 9.59ILAAQRR165 pKa = 11.84QSVLKK170 pKa = 10.1NVVGGAEE177 pKa = 4.16GTEE180 pKa = 3.92ATRR183 pKa = 11.84GQVAQMTYY191 pKa = 10.82NAIDD195 pKa = 3.55TPLMEE200 pKa = 4.49RR201 pKa = 11.84YY202 pKa = 9.9VYY204 pKa = 10.6GGEE207 pKa = 3.85AQYY210 pKa = 10.6VIWDD214 pKa = 3.91GEE216 pKa = 4.33SWSPRR221 pKa = 11.84KK222 pKa = 8.04TLMNQALGINKK233 pKa = 9.66LKK235 pKa = 11.03GVVTEE240 pKa = 4.51NEE242 pKa = 4.07VTALDD247 pKa = 3.76AAVQIDD253 pKa = 3.98TDD255 pKa = 3.46ATQQIKK261 pKa = 10.69LYY263 pKa = 11.18VEE265 pKa = 4.57DD266 pKa = 5.26NYY268 pKa = 11.37LGSNDD273 pKa = 3.64TNSDD277 pKa = 3.88YY278 pKa = 11.16EE279 pKa = 4.17VDD281 pKa = 3.58SVYY284 pKa = 10.79PFYY287 pKa = 10.94TGDD290 pKa = 3.42TNAADD295 pKa = 3.68YY296 pKa = 11.09LGYY299 pKa = 10.82DD300 pKa = 3.57VVLYY304 pKa = 10.79AQDD307 pKa = 3.84NKK309 pKa = 11.18NEE311 pKa = 4.1TDD313 pKa = 3.67TILSITEE320 pKa = 3.84ATGKK324 pKa = 9.75NSKK327 pKa = 10.33VEE329 pKa = 4.03FTLDD333 pKa = 3.07KK334 pKa = 10.83FNSYY338 pKa = 10.85DD339 pKa = 3.59ADD341 pKa = 4.25TNNLSYY347 pKa = 10.48MKK349 pKa = 10.72NDD351 pKa = 3.55TDD353 pKa = 4.8KK354 pKa = 11.49SATKK358 pKa = 10.56LKK360 pKa = 10.78LQTTSNRR367 pKa = 11.84VNYY370 pKa = 9.58SDD372 pKa = 4.31SPAIIYY378 pKa = 10.34NGIAYY383 pKa = 10.3SGTLEE388 pKa = 4.21SLFGSYY394 pKa = 10.23EE395 pKa = 3.91GDD397 pKa = 3.3EE398 pKa = 4.39SGLIYY403 pKa = 10.29KK404 pKa = 10.58DD405 pKa = 3.15SAYY408 pKa = 10.44SGKK411 pKa = 8.64VTVLDD416 pKa = 3.92NDD418 pKa = 3.74DD419 pKa = 3.47TSGYY423 pKa = 10.95DD424 pKa = 3.33VIFVDD429 pKa = 3.94VAVGAVVDD437 pKa = 4.18EE438 pKa = 4.39LSSRR442 pKa = 11.84GVLTFKK448 pKa = 11.03NSVGDD453 pKa = 3.89RR454 pKa = 11.84NHH456 pKa = 6.1MNSVSKK462 pKa = 11.48VEE464 pKa = 4.22FDD466 pKa = 4.0SSNTDD471 pKa = 2.93MYY473 pKa = 11.5INLTKK478 pKa = 10.52DD479 pKa = 3.29GKK481 pKa = 9.92PYY483 pKa = 10.8DD484 pKa = 3.97YY485 pKa = 9.97TALKK489 pKa = 10.02AWDD492 pKa = 3.75VLSIVANNGDD502 pKa = 4.09AQSDD506 pKa = 4.89YY507 pKa = 11.7YY508 pKa = 10.89DD509 pKa = 3.89IKK511 pKa = 11.54VLDD514 pKa = 3.96NTNTAITGSVSRR526 pKa = 11.84TSSSDD531 pKa = 2.89TSADD535 pKa = 2.94GTAYY539 pKa = 9.57TISGVDD545 pKa = 3.24YY546 pKa = 10.89DD547 pKa = 4.58LAEE550 pKa = 4.62GYY552 pKa = 10.51YY553 pKa = 11.12NNGTLKK559 pKa = 10.64AGSEE563 pKa = 4.13GTFYY567 pKa = 10.44IDD569 pKa = 3.22EE570 pKa = 4.34YY571 pKa = 11.29GKK573 pKa = 10.08IVAYY577 pKa = 10.82NKK579 pKa = 10.26DD580 pKa = 4.16SNSKK584 pKa = 8.9TSDD587 pKa = 2.89NYY589 pKa = 11.3GYY591 pKa = 11.06VIDD594 pKa = 4.49AQITSSGFSDD604 pKa = 3.93NVPSLQIIYY613 pKa = 10.33KK614 pKa = 10.1DD615 pKa = 3.81GTIGTYY621 pKa = 10.49DD622 pKa = 3.36FQGTVTIDD630 pKa = 3.23NPTEE634 pKa = 4.3EE635 pKa = 4.17IQKK638 pKa = 10.62LVGDD642 pKa = 4.26NSDD645 pKa = 3.64SVSFKK650 pKa = 11.04YY651 pKa = 10.3KK652 pKa = 10.38DD653 pKa = 3.62YY654 pKa = 11.38SVDD657 pKa = 3.49AMTEE661 pKa = 4.21AINTMAGTVITYY673 pKa = 9.32NATNGYY679 pKa = 9.0IKK681 pKa = 10.36SITMAALNSTTDD693 pKa = 3.18DD694 pKa = 3.51STLVLEE700 pKa = 5.1KK701 pKa = 10.71ADD703 pKa = 3.84TANVSSFDD711 pKa = 3.84EE712 pKa = 4.1DD713 pKa = 3.72TKK715 pKa = 10.62QIRR718 pKa = 11.84VGGTKK723 pKa = 10.02KK724 pKa = 10.75YY725 pKa = 10.95DD726 pKa = 3.41VADD729 pKa = 3.87DD730 pKa = 3.68TLIFYY735 pKa = 10.09IGKK738 pKa = 9.03PGDD741 pKa = 3.47SFVYY745 pKa = 9.97NKK747 pKa = 9.44PAAGEE752 pKa = 3.84PSEE755 pKa = 4.1NCIVGQGSEE764 pKa = 4.76LRR766 pKa = 11.84TNDD769 pKa = 3.33NFSSAVYY776 pKa = 10.47RR777 pKa = 11.84NGDD780 pKa = 3.33EE781 pKa = 4.33SGTASVIVMYY791 pKa = 9.24NTDD794 pKa = 3.85PGVGASTGVAYY805 pKa = 8.63ITSIGQTTVDD815 pKa = 3.36GSKK818 pKa = 10.45VLSITYY824 pKa = 9.63YY825 pKa = 10.27QDD827 pKa = 4.02GEE829 pKa = 4.28LKK831 pKa = 10.37EE832 pKa = 5.29ANTDD836 pKa = 3.53DD837 pKa = 4.15MSVGDD842 pKa = 4.75YY843 pKa = 11.09LDD845 pKa = 3.55QNTVAGSLFKK855 pKa = 10.86FGISNGTITSAVPYY869 pKa = 8.85LTFDD873 pKa = 3.08GTVRR877 pKa = 11.84DD878 pKa = 3.94KK879 pKa = 11.37VYY881 pKa = 10.86EE882 pKa = 4.32GDD884 pKa = 4.0DD885 pKa = 3.59FTTAGIPNANYY896 pKa = 10.51VKK898 pKa = 8.34TTGSNSDD905 pKa = 3.33EE906 pKa = 3.97EE907 pKa = 4.93VYY909 pKa = 11.0YY910 pKa = 10.93GAVVQKK916 pKa = 10.42SGNRR920 pKa = 11.84LTIAQADD927 pKa = 3.56ADD929 pKa = 4.89GIISLDD935 pKa = 3.64PNDD938 pKa = 4.12WEE940 pKa = 4.74SVSLSNYY947 pKa = 5.89TANYY951 pKa = 9.27YY952 pKa = 10.76LYY954 pKa = 10.74DD955 pKa = 3.39PTRR958 pKa = 11.84NGKK961 pKa = 9.81AKK963 pKa = 10.32FQTASLGDD971 pKa = 3.41ITVDD975 pKa = 3.71KK976 pKa = 11.23VLANDD981 pKa = 4.64GGNGSYY987 pKa = 10.34TIDD990 pKa = 3.37FGGVVEE996 pKa = 4.8DD997 pKa = 4.67APAFGMLDD1005 pKa = 3.47FVYY1008 pKa = 10.54VRR1010 pKa = 11.84TYY1012 pKa = 10.86EE1013 pKa = 3.96RR1014 pKa = 11.84TADD1017 pKa = 3.62VVDD1020 pKa = 3.85YY1021 pKa = 7.48TTWEE1025 pKa = 3.67YY1026 pKa = 11.31DD1027 pKa = 3.66YY1028 pKa = 11.39NFSKK1032 pKa = 11.14
MM1 pKa = 7.86RR2 pKa = 11.84NLKK5 pKa = 10.19KK6 pKa = 10.71VIALIAVFAMMVSTVAFAATFGDD29 pKa = 3.96VKK31 pKa = 11.03EE32 pKa = 4.18GDD34 pKa = 3.58NYY36 pKa = 11.26YY37 pKa = 10.78EE38 pKa = 5.57AIEE41 pKa = 4.18TLNKK45 pKa = 10.47LGILTGDD52 pKa = 4.06DD53 pKa = 3.79EE54 pKa = 5.62NNDD57 pKa = 2.72GVMEE61 pKa = 3.84YY62 pKa = 10.36RR63 pKa = 11.84PNDD66 pKa = 3.91SITRR70 pKa = 11.84AEE72 pKa = 3.79ITVIVARR79 pKa = 11.84IQGQTGAVAQADD91 pKa = 4.1TVFTDD96 pKa = 3.55VPSTYY101 pKa = 9.23WASGYY106 pKa = 9.59IASATNQGIINGYY119 pKa = 10.16GDD121 pKa = 3.86GTFGPDD127 pKa = 3.96DD128 pKa = 3.64KK129 pKa = 11.59VLYY132 pKa = 10.29EE133 pKa = 4.67DD134 pKa = 4.02VIKK137 pKa = 10.51MLMEE141 pKa = 3.88TLGYY145 pKa = 10.01KK146 pKa = 10.19PYY148 pKa = 10.43AQNNGGYY155 pKa = 6.11PTGYY159 pKa = 9.59ILAAQRR165 pKa = 11.84QSVLKK170 pKa = 10.1NVVGGAEE177 pKa = 4.16GTEE180 pKa = 3.92ATRR183 pKa = 11.84GQVAQMTYY191 pKa = 10.82NAIDD195 pKa = 3.55TPLMEE200 pKa = 4.49RR201 pKa = 11.84YY202 pKa = 9.9VYY204 pKa = 10.6GGEE207 pKa = 3.85AQYY210 pKa = 10.6VIWDD214 pKa = 3.91GEE216 pKa = 4.33SWSPRR221 pKa = 11.84KK222 pKa = 8.04TLMNQALGINKK233 pKa = 9.66LKK235 pKa = 11.03GVVTEE240 pKa = 4.51NEE242 pKa = 4.07VTALDD247 pKa = 3.76AAVQIDD253 pKa = 3.98TDD255 pKa = 3.46ATQQIKK261 pKa = 10.69LYY263 pKa = 11.18VEE265 pKa = 4.57DD266 pKa = 5.26NYY268 pKa = 11.37LGSNDD273 pKa = 3.64TNSDD277 pKa = 3.88YY278 pKa = 11.16EE279 pKa = 4.17VDD281 pKa = 3.58SVYY284 pKa = 10.79PFYY287 pKa = 10.94TGDD290 pKa = 3.42TNAADD295 pKa = 3.68YY296 pKa = 11.09LGYY299 pKa = 10.82DD300 pKa = 3.57VVLYY304 pKa = 10.79AQDD307 pKa = 3.84NKK309 pKa = 11.18NEE311 pKa = 4.1TDD313 pKa = 3.67TILSITEE320 pKa = 3.84ATGKK324 pKa = 9.75NSKK327 pKa = 10.33VEE329 pKa = 4.03FTLDD333 pKa = 3.07KK334 pKa = 10.83FNSYY338 pKa = 10.85DD339 pKa = 3.59ADD341 pKa = 4.25TNNLSYY347 pKa = 10.48MKK349 pKa = 10.72NDD351 pKa = 3.55TDD353 pKa = 4.8KK354 pKa = 11.49SATKK358 pKa = 10.56LKK360 pKa = 10.78LQTTSNRR367 pKa = 11.84VNYY370 pKa = 9.58SDD372 pKa = 4.31SPAIIYY378 pKa = 10.34NGIAYY383 pKa = 10.3SGTLEE388 pKa = 4.21SLFGSYY394 pKa = 10.23EE395 pKa = 3.91GDD397 pKa = 3.3EE398 pKa = 4.39SGLIYY403 pKa = 10.29KK404 pKa = 10.58DD405 pKa = 3.15SAYY408 pKa = 10.44SGKK411 pKa = 8.64VTVLDD416 pKa = 3.92NDD418 pKa = 3.74DD419 pKa = 3.47TSGYY423 pKa = 10.95DD424 pKa = 3.33VIFVDD429 pKa = 3.94VAVGAVVDD437 pKa = 4.18EE438 pKa = 4.39LSSRR442 pKa = 11.84GVLTFKK448 pKa = 11.03NSVGDD453 pKa = 3.89RR454 pKa = 11.84NHH456 pKa = 6.1MNSVSKK462 pKa = 11.48VEE464 pKa = 4.22FDD466 pKa = 4.0SSNTDD471 pKa = 2.93MYY473 pKa = 11.5INLTKK478 pKa = 10.52DD479 pKa = 3.29GKK481 pKa = 9.92PYY483 pKa = 10.8DD484 pKa = 3.97YY485 pKa = 9.97TALKK489 pKa = 10.02AWDD492 pKa = 3.75VLSIVANNGDD502 pKa = 4.09AQSDD506 pKa = 4.89YY507 pKa = 11.7YY508 pKa = 10.89DD509 pKa = 3.89IKK511 pKa = 11.54VLDD514 pKa = 3.96NTNTAITGSVSRR526 pKa = 11.84TSSSDD531 pKa = 2.89TSADD535 pKa = 2.94GTAYY539 pKa = 9.57TISGVDD545 pKa = 3.24YY546 pKa = 10.89DD547 pKa = 4.58LAEE550 pKa = 4.62GYY552 pKa = 10.51YY553 pKa = 11.12NNGTLKK559 pKa = 10.64AGSEE563 pKa = 4.13GTFYY567 pKa = 10.44IDD569 pKa = 3.22EE570 pKa = 4.34YY571 pKa = 11.29GKK573 pKa = 10.08IVAYY577 pKa = 10.82NKK579 pKa = 10.26DD580 pKa = 4.16SNSKK584 pKa = 8.9TSDD587 pKa = 2.89NYY589 pKa = 11.3GYY591 pKa = 11.06VIDD594 pKa = 4.49AQITSSGFSDD604 pKa = 3.93NVPSLQIIYY613 pKa = 10.33KK614 pKa = 10.1DD615 pKa = 3.81GTIGTYY621 pKa = 10.49DD622 pKa = 3.36FQGTVTIDD630 pKa = 3.23NPTEE634 pKa = 4.3EE635 pKa = 4.17IQKK638 pKa = 10.62LVGDD642 pKa = 4.26NSDD645 pKa = 3.64SVSFKK650 pKa = 11.04YY651 pKa = 10.3KK652 pKa = 10.38DD653 pKa = 3.62YY654 pKa = 11.38SVDD657 pKa = 3.49AMTEE661 pKa = 4.21AINTMAGTVITYY673 pKa = 9.32NATNGYY679 pKa = 9.0IKK681 pKa = 10.36SITMAALNSTTDD693 pKa = 3.18DD694 pKa = 3.51STLVLEE700 pKa = 5.1KK701 pKa = 10.71ADD703 pKa = 3.84TANVSSFDD711 pKa = 3.84EE712 pKa = 4.1DD713 pKa = 3.72TKK715 pKa = 10.62QIRR718 pKa = 11.84VGGTKK723 pKa = 10.02KK724 pKa = 10.75YY725 pKa = 10.95DD726 pKa = 3.41VADD729 pKa = 3.87DD730 pKa = 3.68TLIFYY735 pKa = 10.09IGKK738 pKa = 9.03PGDD741 pKa = 3.47SFVYY745 pKa = 9.97NKK747 pKa = 9.44PAAGEE752 pKa = 3.84PSEE755 pKa = 4.1NCIVGQGSEE764 pKa = 4.76LRR766 pKa = 11.84TNDD769 pKa = 3.33NFSSAVYY776 pKa = 10.47RR777 pKa = 11.84NGDD780 pKa = 3.33EE781 pKa = 4.33SGTASVIVMYY791 pKa = 9.24NTDD794 pKa = 3.85PGVGASTGVAYY805 pKa = 8.63ITSIGQTTVDD815 pKa = 3.36GSKK818 pKa = 10.45VLSITYY824 pKa = 9.63YY825 pKa = 10.27QDD827 pKa = 4.02GEE829 pKa = 4.28LKK831 pKa = 10.37EE832 pKa = 5.29ANTDD836 pKa = 3.53DD837 pKa = 4.15MSVGDD842 pKa = 4.75YY843 pKa = 11.09LDD845 pKa = 3.55QNTVAGSLFKK855 pKa = 10.86FGISNGTITSAVPYY869 pKa = 8.85LTFDD873 pKa = 3.08GTVRR877 pKa = 11.84DD878 pKa = 3.94KK879 pKa = 11.37VYY881 pKa = 10.86EE882 pKa = 4.32GDD884 pKa = 4.0DD885 pKa = 3.59FTTAGIPNANYY896 pKa = 10.51VKK898 pKa = 8.34TTGSNSDD905 pKa = 3.33EE906 pKa = 3.97EE907 pKa = 4.93VYY909 pKa = 11.0YY910 pKa = 10.93GAVVQKK916 pKa = 10.42SGNRR920 pKa = 11.84LTIAQADD927 pKa = 3.56ADD929 pKa = 4.89GIISLDD935 pKa = 3.64PNDD938 pKa = 4.12WEE940 pKa = 4.74SVSLSNYY947 pKa = 5.89TANYY951 pKa = 9.27YY952 pKa = 10.76LYY954 pKa = 10.74DD955 pKa = 3.39PTRR958 pKa = 11.84NGKK961 pKa = 9.81AKK963 pKa = 10.32FQTASLGDD971 pKa = 3.41ITVDD975 pKa = 3.71KK976 pKa = 11.23VLANDD981 pKa = 4.64GGNGSYY987 pKa = 10.34TIDD990 pKa = 3.37FGGVVEE996 pKa = 4.8DD997 pKa = 4.67APAFGMLDD1005 pKa = 3.47FVYY1008 pKa = 10.54VRR1010 pKa = 11.84TYY1012 pKa = 10.86EE1013 pKa = 3.96RR1014 pKa = 11.84TADD1017 pKa = 3.62VVDD1020 pKa = 3.85YY1021 pKa = 7.48TTWEE1025 pKa = 3.67YY1026 pKa = 11.31DD1027 pKa = 3.66YY1028 pKa = 11.39NFSKK1032 pKa = 11.14
Molecular weight: 111.94 kDa
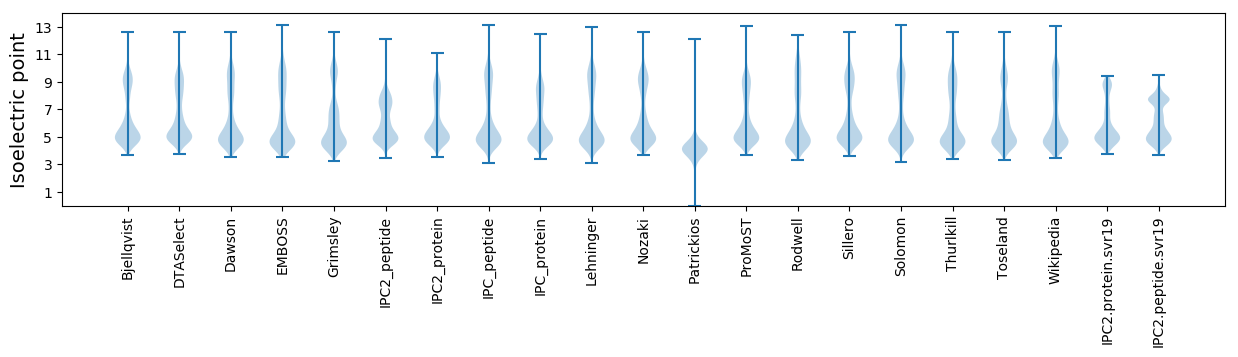
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K9P241|A0A2K9P241_9FIRM Phage protein OS=Monoglobus pectinilyticus OX=1981510 GN=B9O19_00475 PE=4 SV=1
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.82QPKK8 pKa = 9.51KK9 pKa = 7.62RR10 pKa = 11.84HH11 pKa = 5.45RR12 pKa = 11.84SKK14 pKa = 10.49VHH16 pKa = 5.68GFRR19 pKa = 11.84KK20 pKa = 10.02RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.44VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.63GRR39 pKa = 11.84KK40 pKa = 8.9SLSAA44 pKa = 3.86
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.82QPKK8 pKa = 9.51KK9 pKa = 7.62RR10 pKa = 11.84HH11 pKa = 5.45RR12 pKa = 11.84SKK14 pKa = 10.49VHH16 pKa = 5.68GFRR19 pKa = 11.84KK20 pKa = 10.02RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.44VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.63GRR39 pKa = 11.84KK40 pKa = 8.9SLSAA44 pKa = 3.86
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
762331 |
29 |
3174 |
337.6 |
37.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.719 ± 0.053 | 1.385 ± 0.025 |
6.225 ± 0.04 | 7.043 ± 0.054 |
4.046 ± 0.036 | 7.124 ± 0.05 |
1.339 ± 0.018 | 8.157 ± 0.052 |
7.294 ± 0.051 | 7.886 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.634 ± 0.024 | 5.945 ± 0.047 |
3.326 ± 0.039 | 2.611 ± 0.03 |
3.69 ± 0.043 | 6.891 ± 0.053 |
5.741 ± 0.07 | 6.722 ± 0.041 |
0.823 ± 0.019 | 4.398 ± 0.048 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
