
Vulgatibacter incomptus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Cystobacterineae; Vulgatibacteraceae; Vulgatibacter
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
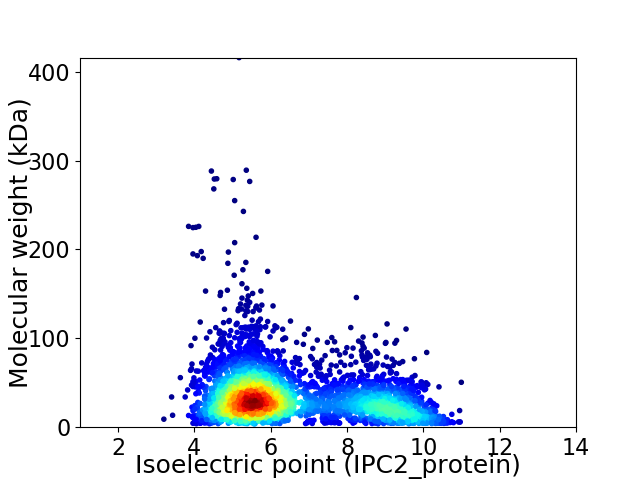
Virtual 2D-PAGE plot for 3704 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1PCL8|A0A0K1PCL8_9DELT ABC transporter related protein OS=Vulgatibacter incomptus OX=1391653 GN=AKJ08_1251 PE=4 SV=1
MM1 pKa = 7.07VLSLGGGCSPSVPDD15 pKa = 4.38GSPAASAGAGGEE27 pKa = 4.39GGGSGGKK34 pKa = 10.19GGGSGGVGGGSGGEE48 pKa = 4.09GGGTGGVGGGTGGVGGGGSTQSPGTHH74 pKa = 6.39FAATARR80 pKa = 11.84EE81 pKa = 4.13WSLPPDD87 pKa = 3.87SPSPDD92 pKa = 2.87GFFTFGYY99 pKa = 10.19YY100 pKa = 10.48DD101 pKa = 4.0WTTMDD106 pKa = 4.65LDD108 pKa = 4.57GDD110 pKa = 4.29GKK112 pKa = 10.06PDD114 pKa = 3.47LVVYY118 pKa = 9.49TGASEE123 pKa = 5.38SNPHH127 pKa = 5.46WLLYY131 pKa = 10.77KK132 pKa = 9.86NTGSGFAATPTEE144 pKa = 4.01WSLPPDD150 pKa = 3.85SPHH153 pKa = 7.01PDD155 pKa = 2.95GFFTLAYY162 pKa = 10.05YY163 pKa = 10.72DD164 pKa = 4.1WTTMDD169 pKa = 4.65LDD171 pKa = 4.57GDD173 pKa = 4.29GKK175 pKa = 10.12PDD177 pKa = 3.56LVVNTGASEE186 pKa = 4.22TNRR189 pKa = 11.84RR190 pKa = 11.84WLDD193 pKa = 3.38YY194 pKa = 10.9KK195 pKa = 10.63NTGTGFAATATEE207 pKa = 4.08WSLPADD213 pKa = 3.91SPHH216 pKa = 7.11PDD218 pKa = 2.93GFFTFSYY225 pKa = 10.79YY226 pKa = 11.07DD227 pKa = 3.63WTTMDD232 pKa = 4.65LDD234 pKa = 4.57GDD236 pKa = 4.29GKK238 pKa = 10.17PDD240 pKa = 3.53LVVNAGASEE249 pKa = 4.24TNRR252 pKa = 11.84RR253 pKa = 11.84WLLYY257 pKa = 10.33KK258 pKa = 9.93NTGSGFAATATEE270 pKa = 4.08WSLPADD276 pKa = 3.66SPFPGGFFTFSYY288 pKa = 10.83YY289 pKa = 11.07DD290 pKa = 3.63WTTMDD295 pKa = 4.65LDD297 pKa = 4.57GDD299 pKa = 4.29GKK301 pKa = 10.06PDD303 pKa = 3.47LVVYY307 pKa = 9.49TGASEE312 pKa = 5.38SNPHH316 pKa = 5.46WLLYY320 pKa = 10.77KK321 pKa = 9.86NTGSGFAATATEE333 pKa = 4.2WSLPPDD339 pKa = 3.76SPSPNGFFTFSYY351 pKa = 10.77YY352 pKa = 11.07DD353 pKa = 3.63WTTMDD358 pKa = 4.65LDD360 pKa = 4.57GDD362 pKa = 4.29GKK364 pKa = 10.06PDD366 pKa = 3.47LVVYY370 pKa = 9.49TGASEE375 pKa = 5.39SNPHH379 pKa = 5.13WLFYY383 pKa = 11.12KK384 pKa = 9.72NTGSGFTATATEE396 pKa = 4.13WTLPADD402 pKa = 4.0SPHH405 pKa = 7.0PDD407 pKa = 2.97GFFTLAYY414 pKa = 10.05YY415 pKa = 10.72DD416 pKa = 4.1WTTMDD421 pKa = 4.65LDD423 pKa = 4.57GDD425 pKa = 4.29GKK427 pKa = 10.12PDD429 pKa = 3.56LVVNTGASEE438 pKa = 4.19TNRR441 pKa = 11.84RR442 pKa = 11.84WLLYY446 pKa = 10.79KK447 pKa = 9.29NTPP450 pKa = 3.29
MM1 pKa = 7.07VLSLGGGCSPSVPDD15 pKa = 4.38GSPAASAGAGGEE27 pKa = 4.39GGGSGGKK34 pKa = 10.19GGGSGGVGGGSGGEE48 pKa = 4.09GGGTGGVGGGTGGVGGGGSTQSPGTHH74 pKa = 6.39FAATARR80 pKa = 11.84EE81 pKa = 4.13WSLPPDD87 pKa = 3.87SPSPDD92 pKa = 2.87GFFTFGYY99 pKa = 10.19YY100 pKa = 10.48DD101 pKa = 4.0WTTMDD106 pKa = 4.65LDD108 pKa = 4.57GDD110 pKa = 4.29GKK112 pKa = 10.06PDD114 pKa = 3.47LVVYY118 pKa = 9.49TGASEE123 pKa = 5.38SNPHH127 pKa = 5.46WLLYY131 pKa = 10.77KK132 pKa = 9.86NTGSGFAATPTEE144 pKa = 4.01WSLPPDD150 pKa = 3.85SPHH153 pKa = 7.01PDD155 pKa = 2.95GFFTLAYY162 pKa = 10.05YY163 pKa = 10.72DD164 pKa = 4.1WTTMDD169 pKa = 4.65LDD171 pKa = 4.57GDD173 pKa = 4.29GKK175 pKa = 10.12PDD177 pKa = 3.56LVVNTGASEE186 pKa = 4.22TNRR189 pKa = 11.84RR190 pKa = 11.84WLDD193 pKa = 3.38YY194 pKa = 10.9KK195 pKa = 10.63NTGTGFAATATEE207 pKa = 4.08WSLPADD213 pKa = 3.91SPHH216 pKa = 7.11PDD218 pKa = 2.93GFFTFSYY225 pKa = 10.79YY226 pKa = 11.07DD227 pKa = 3.63WTTMDD232 pKa = 4.65LDD234 pKa = 4.57GDD236 pKa = 4.29GKK238 pKa = 10.17PDD240 pKa = 3.53LVVNAGASEE249 pKa = 4.24TNRR252 pKa = 11.84RR253 pKa = 11.84WLLYY257 pKa = 10.33KK258 pKa = 9.93NTGSGFAATATEE270 pKa = 4.08WSLPADD276 pKa = 3.66SPFPGGFFTFSYY288 pKa = 10.83YY289 pKa = 11.07DD290 pKa = 3.63WTTMDD295 pKa = 4.65LDD297 pKa = 4.57GDD299 pKa = 4.29GKK301 pKa = 10.06PDD303 pKa = 3.47LVVYY307 pKa = 9.49TGASEE312 pKa = 5.38SNPHH316 pKa = 5.46WLLYY320 pKa = 10.77KK321 pKa = 9.86NTGSGFAATATEE333 pKa = 4.2WSLPPDD339 pKa = 3.76SPSPNGFFTFSYY351 pKa = 10.77YY352 pKa = 11.07DD353 pKa = 3.63WTTMDD358 pKa = 4.65LDD360 pKa = 4.57GDD362 pKa = 4.29GKK364 pKa = 10.06PDD366 pKa = 3.47LVVYY370 pKa = 9.49TGASEE375 pKa = 5.39SNPHH379 pKa = 5.13WLFYY383 pKa = 11.12KK384 pKa = 9.72NTGSGFTATATEE396 pKa = 4.13WTLPADD402 pKa = 4.0SPHH405 pKa = 7.0PDD407 pKa = 2.97GFFTLAYY414 pKa = 10.05YY415 pKa = 10.72DD416 pKa = 4.1WTTMDD421 pKa = 4.65LDD423 pKa = 4.57GDD425 pKa = 4.29GKK427 pKa = 10.12PDD429 pKa = 3.56LVVNTGASEE438 pKa = 4.19TNRR441 pKa = 11.84RR442 pKa = 11.84WLLYY446 pKa = 10.79KK447 pKa = 9.29NTPP450 pKa = 3.29
Molecular weight: 47.36 kDa
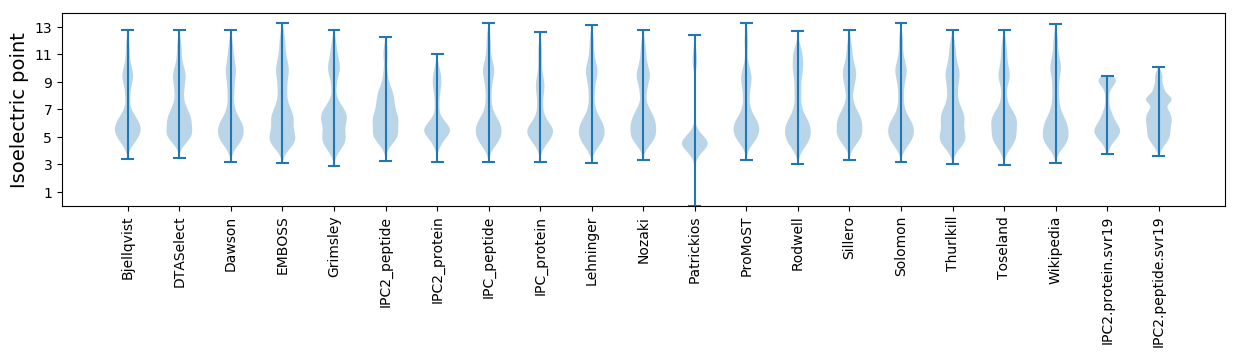
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1PCI4|A0A0K1PCI4_9DELT Acyl carrier protein OS=Vulgatibacter incomptus OX=1391653 GN=acpP PE=3 SV=1
MM1 pKa = 7.44PVVSGSSVRR10 pKa = 11.84ASGRR14 pKa = 11.84GPAASAPGVTASGRR28 pKa = 11.84VRR30 pKa = 11.84AASDD34 pKa = 3.06RR35 pKa = 11.84RR36 pKa = 11.84ATATVRR42 pKa = 11.84DD43 pKa = 4.28RR44 pKa = 11.84TASAPGATATGRR56 pKa = 11.84ARR58 pKa = 11.84TASAPGATAIGRR70 pKa = 11.84GLAASGLRR78 pKa = 11.84ATAIGPALAASVRR91 pKa = 11.84RR92 pKa = 11.84ATATGRR98 pKa = 11.84ARR100 pKa = 11.84TALARR105 pKa = 11.84RR106 pKa = 11.84ATAIGRR112 pKa = 11.84ARR114 pKa = 11.84TALARR119 pKa = 11.84RR120 pKa = 11.84ATAIGRR126 pKa = 11.84ARR128 pKa = 11.84TALARR133 pKa = 11.84RR134 pKa = 11.84ATAIGRR140 pKa = 11.84GPADD144 pKa = 3.09SDD146 pKa = 3.91LRR148 pKa = 11.84VTAIDD153 pKa = 3.54RR154 pKa = 11.84ALAASDD160 pKa = 3.87RR161 pKa = 11.84PAAAIDD167 pKa = 3.62RR168 pKa = 11.84GPAVSDD174 pKa = 3.71PRR176 pKa = 11.84ATAIGRR182 pKa = 11.84ALAASDD188 pKa = 4.39LRR190 pKa = 11.84ATAIGRR196 pKa = 11.84ALAASDD202 pKa = 4.11LRR204 pKa = 11.84ASAIGRR210 pKa = 11.84ARR212 pKa = 11.84TAWARR217 pKa = 11.84RR218 pKa = 11.84ATAIGRR224 pKa = 11.84ALAASDD230 pKa = 4.39LRR232 pKa = 11.84ATAIGRR238 pKa = 11.84APAALDD244 pKa = 3.68LRR246 pKa = 11.84AMAIGRR252 pKa = 11.84ARR254 pKa = 11.84TASARR259 pKa = 11.84RR260 pKa = 11.84AMAIGLARR268 pKa = 11.84TASARR273 pKa = 11.84RR274 pKa = 11.84AMAIGRR280 pKa = 11.84ARR282 pKa = 11.84TASARR287 pKa = 11.84RR288 pKa = 11.84AMAIGLARR296 pKa = 11.84TASARR301 pKa = 11.84RR302 pKa = 11.84ATAIGRR308 pKa = 11.84ARR310 pKa = 11.84TASARR315 pKa = 11.84RR316 pKa = 11.84AMAIGRR322 pKa = 11.84ARR324 pKa = 11.84TALARR329 pKa = 11.84RR330 pKa = 11.84ATAIGRR336 pKa = 11.84ARR338 pKa = 11.84AASAPRR344 pKa = 11.84ATAIGRR350 pKa = 11.84APTASAPRR358 pKa = 11.84AMAIGRR364 pKa = 11.84ARR366 pKa = 11.84TASARR371 pKa = 11.84RR372 pKa = 11.84AMAIGRR378 pKa = 11.84ARR380 pKa = 11.84TALVRR385 pKa = 11.84RR386 pKa = 11.84ATAIGRR392 pKa = 11.84ARR394 pKa = 11.84AASVRR399 pKa = 11.84RR400 pKa = 11.84AMAIGRR406 pKa = 11.84ARR408 pKa = 11.84AASVRR413 pKa = 11.84RR414 pKa = 11.84AMAIGRR420 pKa = 11.84ARR422 pKa = 11.84ATSARR427 pKa = 11.84RR428 pKa = 11.84AMAIVRR434 pKa = 11.84ARR436 pKa = 11.84TASARR441 pKa = 11.84RR442 pKa = 11.84ATAIGRR448 pKa = 11.84ARR450 pKa = 11.84TASAPDD456 pKa = 3.42ATATAPIAASIGRR469 pKa = 11.84LVPARR474 pKa = 11.84PAMPRR479 pKa = 11.84GGLRR483 pKa = 11.84AAPIGVRR490 pKa = 11.84RR491 pKa = 11.84PGAAEE496 pKa = 3.64ATIGRR501 pKa = 11.84VAA503 pKa = 3.74
MM1 pKa = 7.44PVVSGSSVRR10 pKa = 11.84ASGRR14 pKa = 11.84GPAASAPGVTASGRR28 pKa = 11.84VRR30 pKa = 11.84AASDD34 pKa = 3.06RR35 pKa = 11.84RR36 pKa = 11.84ATATVRR42 pKa = 11.84DD43 pKa = 4.28RR44 pKa = 11.84TASAPGATATGRR56 pKa = 11.84ARR58 pKa = 11.84TASAPGATAIGRR70 pKa = 11.84GLAASGLRR78 pKa = 11.84ATAIGPALAASVRR91 pKa = 11.84RR92 pKa = 11.84ATATGRR98 pKa = 11.84ARR100 pKa = 11.84TALARR105 pKa = 11.84RR106 pKa = 11.84ATAIGRR112 pKa = 11.84ARR114 pKa = 11.84TALARR119 pKa = 11.84RR120 pKa = 11.84ATAIGRR126 pKa = 11.84ARR128 pKa = 11.84TALARR133 pKa = 11.84RR134 pKa = 11.84ATAIGRR140 pKa = 11.84GPADD144 pKa = 3.09SDD146 pKa = 3.91LRR148 pKa = 11.84VTAIDD153 pKa = 3.54RR154 pKa = 11.84ALAASDD160 pKa = 3.87RR161 pKa = 11.84PAAAIDD167 pKa = 3.62RR168 pKa = 11.84GPAVSDD174 pKa = 3.71PRR176 pKa = 11.84ATAIGRR182 pKa = 11.84ALAASDD188 pKa = 4.39LRR190 pKa = 11.84ATAIGRR196 pKa = 11.84ALAASDD202 pKa = 4.11LRR204 pKa = 11.84ASAIGRR210 pKa = 11.84ARR212 pKa = 11.84TAWARR217 pKa = 11.84RR218 pKa = 11.84ATAIGRR224 pKa = 11.84ALAASDD230 pKa = 4.39LRR232 pKa = 11.84ATAIGRR238 pKa = 11.84APAALDD244 pKa = 3.68LRR246 pKa = 11.84AMAIGRR252 pKa = 11.84ARR254 pKa = 11.84TASARR259 pKa = 11.84RR260 pKa = 11.84AMAIGLARR268 pKa = 11.84TASARR273 pKa = 11.84RR274 pKa = 11.84AMAIGRR280 pKa = 11.84ARR282 pKa = 11.84TASARR287 pKa = 11.84RR288 pKa = 11.84AMAIGLARR296 pKa = 11.84TASARR301 pKa = 11.84RR302 pKa = 11.84ATAIGRR308 pKa = 11.84ARR310 pKa = 11.84TASARR315 pKa = 11.84RR316 pKa = 11.84AMAIGRR322 pKa = 11.84ARR324 pKa = 11.84TALARR329 pKa = 11.84RR330 pKa = 11.84ATAIGRR336 pKa = 11.84ARR338 pKa = 11.84AASAPRR344 pKa = 11.84ATAIGRR350 pKa = 11.84APTASAPRR358 pKa = 11.84AMAIGRR364 pKa = 11.84ARR366 pKa = 11.84TASARR371 pKa = 11.84RR372 pKa = 11.84AMAIGRR378 pKa = 11.84ARR380 pKa = 11.84TALVRR385 pKa = 11.84RR386 pKa = 11.84ATAIGRR392 pKa = 11.84ARR394 pKa = 11.84AASVRR399 pKa = 11.84RR400 pKa = 11.84AMAIGRR406 pKa = 11.84ARR408 pKa = 11.84AASVRR413 pKa = 11.84RR414 pKa = 11.84AMAIGRR420 pKa = 11.84ARR422 pKa = 11.84ATSARR427 pKa = 11.84RR428 pKa = 11.84AMAIVRR434 pKa = 11.84ARR436 pKa = 11.84TASARR441 pKa = 11.84RR442 pKa = 11.84ATAIGRR448 pKa = 11.84ARR450 pKa = 11.84TASAPDD456 pKa = 3.42ATATAPIAASIGRR469 pKa = 11.84LVPARR474 pKa = 11.84PAMPRR479 pKa = 11.84GGLRR483 pKa = 11.84AAPIGVRR490 pKa = 11.84RR491 pKa = 11.84PGAAEE496 pKa = 3.64ATIGRR501 pKa = 11.84VAA503 pKa = 3.74
Molecular weight: 50.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1270377 |
37 |
3785 |
343.0 |
37.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.517 ± 0.066 | 1.036 ± 0.029 |
5.584 ± 0.032 | 6.777 ± 0.046 |
3.486 ± 0.023 | 9.322 ± 0.056 |
1.884 ± 0.018 | 4.02 ± 0.029 |
2.998 ± 0.039 | 10.398 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.761 ± 0.019 | 1.99 ± 0.024 |
5.714 ± 0.033 | 2.607 ± 0.022 |
8.173 ± 0.05 | 5.927 ± 0.042 |
4.831 ± 0.056 | 7.753 ± 0.037 |
1.32 ± 0.018 | 1.902 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
