
Paenimaribius caenipelagi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paenimaribius
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
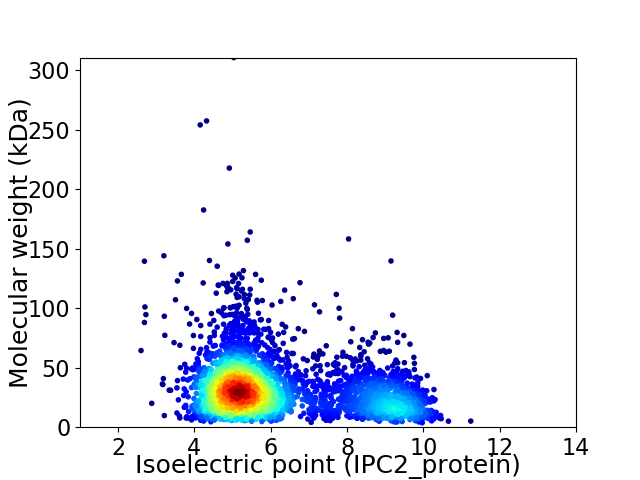
Virtual 2D-PAGE plot for 3702 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
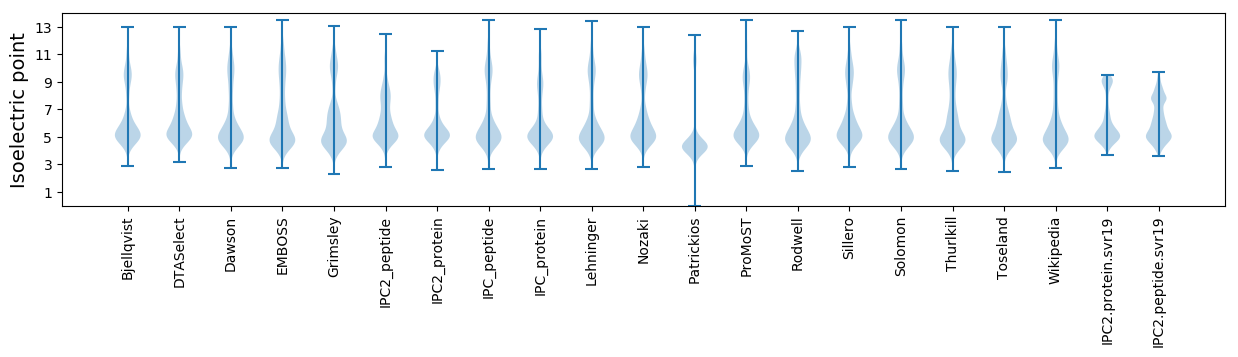
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A547PIS5|A0A547PIS5_9RHOB Uncharacterized protein OS=Paenimaribius caenipelagi OX=2489174 GN=FEV53_19610 PE=4 SV=1
MM1 pKa = 7.85AEE3 pKa = 4.77PDD5 pKa = 3.62TEE7 pKa = 4.3LTVPLQEE14 pKa = 4.92QSDD17 pKa = 4.0LLYY20 pKa = 10.79FDD22 pKa = 5.77FNLDD26 pKa = 3.32GVEE29 pKa = 5.12DD30 pKa = 4.54IIQIRR35 pKa = 11.84RR36 pKa = 11.84SDD38 pKa = 4.02SVWTGTQFLDD48 pKa = 2.79IWSVEE53 pKa = 3.89FWEE56 pKa = 4.69VGSLSDD62 pKa = 4.89PGFSHH67 pKa = 7.62PDD69 pKa = 3.08DD70 pKa = 3.81VAILHH75 pKa = 6.36PVTVEE80 pKa = 3.66AVKK83 pKa = 10.9YY84 pKa = 9.87LASADD89 pKa = 3.97FNKK92 pKa = 10.68DD93 pKa = 2.53LYY95 pKa = 11.43NDD97 pKa = 3.32VKK99 pKa = 10.83RR100 pKa = 11.84PHH102 pKa = 7.5DD103 pKa = 3.78YY104 pKa = 11.31AQTLTLADD112 pKa = 4.56DD113 pKa = 4.7LNSDD117 pKa = 4.48GYY119 pKa = 11.85VDD121 pKa = 4.18LQFSVGFNYY130 pKa = 10.0EE131 pKa = 3.92DD132 pKa = 3.71PLYY135 pKa = 10.56GSRR138 pKa = 11.84YY139 pKa = 9.84EE140 pKa = 4.0YY141 pKa = 10.02LTEE144 pKa = 4.29NGIEE148 pKa = 3.97FAGYY152 pKa = 9.66ILDD155 pKa = 3.6NGRR158 pKa = 11.84YY159 pKa = 8.48QDD161 pKa = 3.87HH162 pKa = 6.99TDD164 pKa = 4.2DD165 pKa = 5.05FYY167 pKa = 11.85GDD169 pKa = 4.08PPHH172 pKa = 6.36QLSEE176 pKa = 4.21IIQAFEE182 pKa = 3.91RR183 pKa = 11.84VISGSVDD190 pKa = 3.57LDD192 pKa = 3.43EE193 pKa = 7.21DD194 pKa = 3.94GALEE198 pKa = 4.11KK199 pKa = 10.52FAVLDD204 pKa = 3.87FQSRR208 pKa = 11.84SWGSLYY214 pKa = 11.0DD215 pKa = 3.73GDD217 pKa = 4.89SNIIEE222 pKa = 4.18SWDD225 pKa = 3.61RR226 pKa = 11.84QQSANYY232 pKa = 9.1VEE234 pKa = 5.82LYY236 pKa = 10.84EE237 pKa = 5.99FDD239 pKa = 3.95DD240 pKa = 4.67QLAIWRR246 pKa = 11.84DD247 pKa = 3.39SEE249 pKa = 4.84EE250 pKa = 4.31YY251 pKa = 10.33PNTPVPLAYY260 pKa = 10.61DD261 pKa = 3.95NVWGLIPTLIFDD273 pKa = 4.63SDD275 pKa = 3.89DD276 pKa = 3.85PLNVEE281 pKa = 4.39QVMSTLSRR289 pKa = 11.84LPGYY293 pKa = 7.58GTQGFYY299 pKa = 10.86EE300 pKa = 4.66GLSSYY305 pKa = 11.03DD306 pKa = 2.99WYY308 pKa = 9.8TAYY311 pKa = 10.5HH312 pKa = 6.76DD313 pKa = 4.43WYY315 pKa = 8.69TGPGGSVDD323 pKa = 3.75YY324 pKa = 11.29DD325 pKa = 4.05AMFLFANSINANQDD339 pKa = 2.88PTFQLGMTQALLQDD353 pKa = 4.23ALDD356 pKa = 3.81SRR358 pKa = 11.84GGSISVDD365 pKa = 3.21LDD367 pKa = 3.54EE368 pKa = 6.81FITDD372 pKa = 4.38PDD374 pKa = 3.87PTGDD378 pKa = 3.85PLTYY382 pKa = 10.46SAIDD386 pKa = 3.8LPAGLILDD394 pKa = 3.98SSTGVVSGASEE405 pKa = 4.05EE406 pKa = 4.49TMTTSFMADD415 pKa = 3.47DD416 pKa = 4.12GRR418 pKa = 11.84GGTFTTTVSFSLPGLDD434 pKa = 4.74GIVDD438 pKa = 4.26GTAGGDD444 pKa = 3.49VITTNFVDD452 pKa = 4.64AYY454 pKa = 10.46GDD456 pKa = 3.39TLTFSKK462 pKa = 10.65VDD464 pKa = 3.59HH465 pKa = 6.58KK466 pKa = 11.61VLGYY470 pKa = 10.77EE471 pKa = 3.87GDD473 pKa = 3.93DD474 pKa = 3.69YY475 pKa = 11.66IGIAKK480 pKa = 10.32GIGIEE485 pKa = 4.11VSGGAGNDD493 pKa = 3.37VIRR496 pKa = 11.84INSATGTASGDD507 pKa = 3.5GGNDD511 pKa = 3.67TIHH514 pKa = 6.19TGVRR518 pKa = 11.84SMVVDD523 pKa = 4.28GGTGDD528 pKa = 4.93DD529 pKa = 3.42MLIANMCYY537 pKa = 9.81GAKK540 pKa = 8.47HH541 pKa = 5.68TLTGGEE547 pKa = 4.17GADD550 pKa = 3.16TFVFTAASTRR560 pKa = 11.84KK561 pKa = 8.97MSVATVTDD569 pKa = 4.38FDD571 pKa = 4.9PEE573 pKa = 4.52LDD575 pKa = 3.57TLVIEE580 pKa = 4.8GEE582 pKa = 4.42VVDD585 pKa = 6.55LSRR588 pKa = 11.84AWLYY592 pKa = 10.86DD593 pKa = 3.17FRR595 pKa = 11.84SITTTAGGDD604 pKa = 3.23RR605 pKa = 11.84VLGYY609 pKa = 10.82GDD611 pKa = 3.56NDD613 pKa = 3.91EE614 pKa = 4.92IVISNLIDD622 pKa = 3.57VDD624 pKa = 3.84DD625 pKa = 5.26LLYY628 pKa = 10.91QFF630 pKa = 5.61
MM1 pKa = 7.85AEE3 pKa = 4.77PDD5 pKa = 3.62TEE7 pKa = 4.3LTVPLQEE14 pKa = 4.92QSDD17 pKa = 4.0LLYY20 pKa = 10.79FDD22 pKa = 5.77FNLDD26 pKa = 3.32GVEE29 pKa = 5.12DD30 pKa = 4.54IIQIRR35 pKa = 11.84RR36 pKa = 11.84SDD38 pKa = 4.02SVWTGTQFLDD48 pKa = 2.79IWSVEE53 pKa = 3.89FWEE56 pKa = 4.69VGSLSDD62 pKa = 4.89PGFSHH67 pKa = 7.62PDD69 pKa = 3.08DD70 pKa = 3.81VAILHH75 pKa = 6.36PVTVEE80 pKa = 3.66AVKK83 pKa = 10.9YY84 pKa = 9.87LASADD89 pKa = 3.97FNKK92 pKa = 10.68DD93 pKa = 2.53LYY95 pKa = 11.43NDD97 pKa = 3.32VKK99 pKa = 10.83RR100 pKa = 11.84PHH102 pKa = 7.5DD103 pKa = 3.78YY104 pKa = 11.31AQTLTLADD112 pKa = 4.56DD113 pKa = 4.7LNSDD117 pKa = 4.48GYY119 pKa = 11.85VDD121 pKa = 4.18LQFSVGFNYY130 pKa = 10.0EE131 pKa = 3.92DD132 pKa = 3.71PLYY135 pKa = 10.56GSRR138 pKa = 11.84YY139 pKa = 9.84EE140 pKa = 4.0YY141 pKa = 10.02LTEE144 pKa = 4.29NGIEE148 pKa = 3.97FAGYY152 pKa = 9.66ILDD155 pKa = 3.6NGRR158 pKa = 11.84YY159 pKa = 8.48QDD161 pKa = 3.87HH162 pKa = 6.99TDD164 pKa = 4.2DD165 pKa = 5.05FYY167 pKa = 11.85GDD169 pKa = 4.08PPHH172 pKa = 6.36QLSEE176 pKa = 4.21IIQAFEE182 pKa = 3.91RR183 pKa = 11.84VISGSVDD190 pKa = 3.57LDD192 pKa = 3.43EE193 pKa = 7.21DD194 pKa = 3.94GALEE198 pKa = 4.11KK199 pKa = 10.52FAVLDD204 pKa = 3.87FQSRR208 pKa = 11.84SWGSLYY214 pKa = 11.0DD215 pKa = 3.73GDD217 pKa = 4.89SNIIEE222 pKa = 4.18SWDD225 pKa = 3.61RR226 pKa = 11.84QQSANYY232 pKa = 9.1VEE234 pKa = 5.82LYY236 pKa = 10.84EE237 pKa = 5.99FDD239 pKa = 3.95DD240 pKa = 4.67QLAIWRR246 pKa = 11.84DD247 pKa = 3.39SEE249 pKa = 4.84EE250 pKa = 4.31YY251 pKa = 10.33PNTPVPLAYY260 pKa = 10.61DD261 pKa = 3.95NVWGLIPTLIFDD273 pKa = 4.63SDD275 pKa = 3.89DD276 pKa = 3.85PLNVEE281 pKa = 4.39QVMSTLSRR289 pKa = 11.84LPGYY293 pKa = 7.58GTQGFYY299 pKa = 10.86EE300 pKa = 4.66GLSSYY305 pKa = 11.03DD306 pKa = 2.99WYY308 pKa = 9.8TAYY311 pKa = 10.5HH312 pKa = 6.76DD313 pKa = 4.43WYY315 pKa = 8.69TGPGGSVDD323 pKa = 3.75YY324 pKa = 11.29DD325 pKa = 4.05AMFLFANSINANQDD339 pKa = 2.88PTFQLGMTQALLQDD353 pKa = 4.23ALDD356 pKa = 3.81SRR358 pKa = 11.84GGSISVDD365 pKa = 3.21LDD367 pKa = 3.54EE368 pKa = 6.81FITDD372 pKa = 4.38PDD374 pKa = 3.87PTGDD378 pKa = 3.85PLTYY382 pKa = 10.46SAIDD386 pKa = 3.8LPAGLILDD394 pKa = 3.98SSTGVVSGASEE405 pKa = 4.05EE406 pKa = 4.49TMTTSFMADD415 pKa = 3.47DD416 pKa = 4.12GRR418 pKa = 11.84GGTFTTTVSFSLPGLDD434 pKa = 4.74GIVDD438 pKa = 4.26GTAGGDD444 pKa = 3.49VITTNFVDD452 pKa = 4.64AYY454 pKa = 10.46GDD456 pKa = 3.39TLTFSKK462 pKa = 10.65VDD464 pKa = 3.59HH465 pKa = 6.58KK466 pKa = 11.61VLGYY470 pKa = 10.77EE471 pKa = 3.87GDD473 pKa = 3.93DD474 pKa = 3.69YY475 pKa = 11.66IGIAKK480 pKa = 10.32GIGIEE485 pKa = 4.11VSGGAGNDD493 pKa = 3.37VIRR496 pKa = 11.84INSATGTASGDD507 pKa = 3.5GGNDD511 pKa = 3.67TIHH514 pKa = 6.19TGVRR518 pKa = 11.84SMVVDD523 pKa = 4.28GGTGDD528 pKa = 4.93DD529 pKa = 3.42MLIANMCYY537 pKa = 9.81GAKK540 pKa = 8.47HH541 pKa = 5.68TLTGGEE547 pKa = 4.17GADD550 pKa = 3.16TFVFTAASTRR560 pKa = 11.84KK561 pKa = 8.97MSVATVTDD569 pKa = 4.38FDD571 pKa = 4.9PEE573 pKa = 4.52LDD575 pKa = 3.57TLVIEE580 pKa = 4.8GEE582 pKa = 4.42VVDD585 pKa = 6.55LSRR588 pKa = 11.84AWLYY592 pKa = 10.86DD593 pKa = 3.17FRR595 pKa = 11.84SITTTAGGDD604 pKa = 3.23RR605 pKa = 11.84VLGYY609 pKa = 10.82GDD611 pKa = 3.56NDD613 pKa = 3.91EE614 pKa = 4.92IVISNLIDD622 pKa = 3.57VDD624 pKa = 3.84DD625 pKa = 5.26LLYY628 pKa = 10.91QFF630 pKa = 5.61
Molecular weight: 68.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A547Q757|A0A547Q757_9RHOB MBL fold metallo-hydrolase OS=Paenimaribius caenipelagi OX=2489174 GN=FEV53_05705 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1141485 |
32 |
2956 |
308.3 |
33.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.803 ± 0.05 | 0.937 ± 0.017 |
6.268 ± 0.045 | 6.119 ± 0.041 |
3.578 ± 0.027 | 8.763 ± 0.059 |
2.054 ± 0.023 | 5.176 ± 0.028 |
2.99 ± 0.032 | 10.04 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.603 ± 0.021 | 2.553 ± 0.024 |
5.096 ± 0.031 | 3.155 ± 0.02 |
7.152 ± 0.048 | 5.392 ± 0.029 |
5.574 ± 0.031 | 7.137 ± 0.037 |
1.398 ± 0.015 | 2.211 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
