
Eel River basin pequenovirus
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
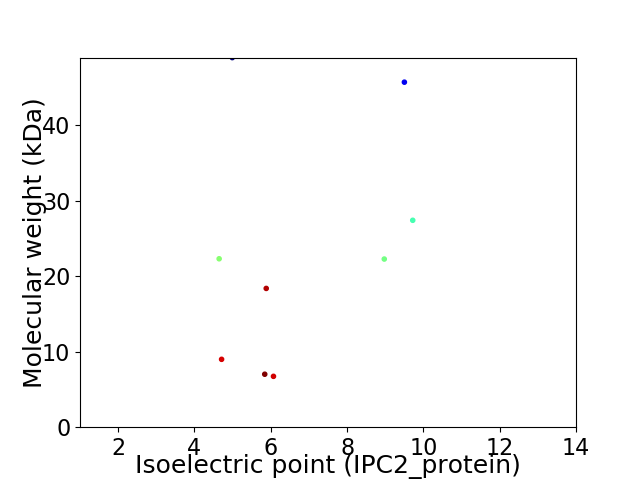
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
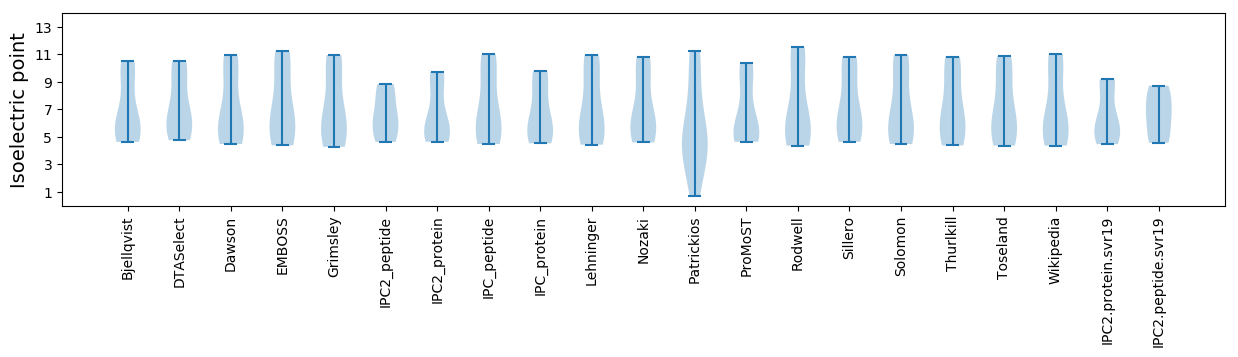
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5AK08|A0A0C5AK08_9VIRU Putative replication initiation protein OS=Eel River basin pequenovirus OX=1609634 PE=4 SV=1
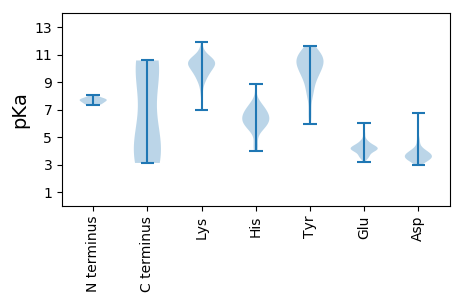
MM1 pKa = 7.59LVPEE5 pKa = 4.25VQGNQLYY12 pKa = 10.77YY13 pKa = 10.56DD14 pKa = 3.9ISSKK18 pKa = 9.62TEE20 pKa = 3.75FSTSEE25 pKa = 3.39IAAIDD30 pKa = 3.88SNISGDD36 pKa = 3.45EE37 pKa = 4.14YY38 pKa = 11.58YY39 pKa = 10.19PYY41 pKa = 11.41AWTSTVNDD49 pKa = 3.19EE50 pKa = 4.23LANYY54 pKa = 9.69RR55 pKa = 11.84YY56 pKa = 10.18YY57 pKa = 11.33VDD59 pKa = 3.84NSVDD63 pKa = 3.61LVAGSRR69 pKa = 11.84VAFGIFLAPDD79 pKa = 3.45VNKK82 pKa = 11.07DD83 pKa = 3.14NMLINIEE90 pKa = 3.99GAMAFYY96 pKa = 10.93GDD98 pKa = 3.54STRR101 pKa = 11.84FNVYY105 pKa = 9.77PILGTTVSSTITSSKK120 pKa = 10.2AAQSNQLACWKK131 pKa = 9.71VIPCEE136 pKa = 4.19TQDD139 pKa = 3.28GAGYY143 pKa = 10.27SKK145 pKa = 10.76LVSFNKK151 pKa = 9.22TVLAVEE157 pKa = 4.09QSSVNTYY164 pKa = 10.02CLAWVISNHH173 pKa = 5.71EE174 pKa = 3.95GSTRR178 pKa = 11.84SLEE181 pKa = 4.31FNSNLSMRR189 pKa = 11.84AWTTPINYY197 pKa = 8.24HH198 pKa = 6.43RR199 pKa = 11.84PGAA202 pKa = 3.85
MM1 pKa = 7.59LVPEE5 pKa = 4.25VQGNQLYY12 pKa = 10.77YY13 pKa = 10.56DD14 pKa = 3.9ISSKK18 pKa = 9.62TEE20 pKa = 3.75FSTSEE25 pKa = 3.39IAAIDD30 pKa = 3.88SNISGDD36 pKa = 3.45EE37 pKa = 4.14YY38 pKa = 11.58YY39 pKa = 10.19PYY41 pKa = 11.41AWTSTVNDD49 pKa = 3.19EE50 pKa = 4.23LANYY54 pKa = 9.69RR55 pKa = 11.84YY56 pKa = 10.18YY57 pKa = 11.33VDD59 pKa = 3.84NSVDD63 pKa = 3.61LVAGSRR69 pKa = 11.84VAFGIFLAPDD79 pKa = 3.45VNKK82 pKa = 11.07DD83 pKa = 3.14NMLINIEE90 pKa = 3.99GAMAFYY96 pKa = 10.93GDD98 pKa = 3.54STRR101 pKa = 11.84FNVYY105 pKa = 9.77PILGTTVSSTITSSKK120 pKa = 10.2AAQSNQLACWKK131 pKa = 9.71VIPCEE136 pKa = 4.19TQDD139 pKa = 3.28GAGYY143 pKa = 10.27SKK145 pKa = 10.76LVSFNKK151 pKa = 9.22TVLAVEE157 pKa = 4.09QSSVNTYY164 pKa = 10.02CLAWVISNHH173 pKa = 5.71EE174 pKa = 3.95GSTRR178 pKa = 11.84SLEE181 pKa = 4.31FNSNLSMRR189 pKa = 11.84AWTTPINYY197 pKa = 8.24HH198 pKa = 6.43RR199 pKa = 11.84PGAA202 pKa = 3.85
Molecular weight: 22.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5AK13|A0A0C5AK13_9VIRU Uncharacterized protein OS=Eel River basin pequenovirus OX=1609634 PE=4 SV=1
MM1 pKa = 7.53LSIIKK6 pKa = 9.28NQKK9 pKa = 8.98LKK11 pKa = 11.05KK12 pKa = 10.44LIIRR16 pKa = 11.84YY17 pKa = 9.47KK18 pKa = 9.5MRR20 pKa = 11.84SLRR23 pKa = 11.84LSISKK28 pKa = 10.02SLKK31 pKa = 9.13NLKK34 pKa = 9.92EE35 pKa = 3.95NSLKK39 pKa = 10.54ARR41 pKa = 11.84KK42 pKa = 9.66AEE44 pKa = 3.85LLTRR48 pKa = 11.84LEE50 pKa = 4.16YY51 pKa = 10.54EE52 pKa = 3.6LEE54 pKa = 3.99YY55 pKa = 10.46RR56 pKa = 11.84IKK58 pKa = 10.93NGWYY62 pKa = 10.31CIFNTLTVNEE72 pKa = 4.17SSIEE76 pKa = 4.0KK77 pKa = 10.1VFEE80 pKa = 4.12KK81 pKa = 10.62GSRR84 pKa = 11.84IFSDD88 pKa = 3.72YY89 pKa = 10.69VRR91 pKa = 11.84SLDD94 pKa = 3.56RR95 pKa = 11.84GVGIAIHH102 pKa = 6.12KK103 pKa = 9.13NWRR106 pKa = 11.84QAVTKK111 pKa = 9.95RR112 pKa = 11.84KK113 pKa = 9.46EE114 pKa = 3.96GNEE117 pKa = 3.51FHH119 pKa = 6.96TYY121 pKa = 9.02FAVVEE126 pKa = 4.24RR127 pKa = 11.84GTKK130 pKa = 9.63NGRR133 pKa = 11.84LHH135 pKa = 6.1IHH137 pKa = 6.84VIHH140 pKa = 6.3MMKK143 pKa = 10.12EE144 pKa = 4.08LPNGCVDD151 pKa = 4.06PNAGRR156 pKa = 11.84AIPNRR161 pKa = 11.84RR162 pKa = 11.84EE163 pKa = 3.44VTYY166 pKa = 10.56LKK168 pKa = 10.42RR169 pKa = 11.84YY170 pKa = 6.75WKK172 pKa = 10.01YY173 pKa = 10.93GYY175 pKa = 9.36SAPIAVRR182 pKa = 11.84FNTNDD187 pKa = 3.29AFGKK191 pKa = 10.08KK192 pKa = 6.95YY193 pKa = 9.55WRR195 pKa = 11.84FPVKK199 pKa = 10.06EE200 pKa = 3.83VAKK203 pKa = 10.83NRR205 pKa = 11.84FEE207 pKa = 4.65SLEE210 pKa = 4.22CKK212 pKa = 10.28DD213 pKa = 3.49AGSIIGYY220 pKa = 8.39IGKK223 pKa = 10.04YY224 pKa = 5.93MTKK227 pKa = 10.05EE228 pKa = 3.48YY229 pKa = 10.75DD230 pKa = 3.26KK231 pKa = 10.45PLEE234 pKa = 4.18KK235 pKa = 10.35EE236 pKa = 4.05YY237 pKa = 10.0VTWRR241 pKa = 11.84TRR243 pKa = 11.84LTRR246 pKa = 11.84TLGTQIMKK254 pKa = 9.99SLCRR258 pKa = 11.84VVTKK262 pKa = 10.85NQLLLLMTMKK272 pKa = 9.38STKK275 pKa = 8.21EE276 pKa = 3.68MKK278 pKa = 10.43VLEE281 pKa = 4.76KK282 pKa = 10.39PLPLYY287 pKa = 10.45KK288 pKa = 10.43LRR290 pKa = 11.84TIATQEE296 pKa = 3.65YY297 pKa = 8.27MRR299 pKa = 11.84RR300 pKa = 11.84SLTNQPLKK308 pKa = 10.16IWEE311 pKa = 4.32SLICLTPRR319 pKa = 11.84MSLLTHH325 pKa = 6.05VKK327 pKa = 10.84SMIQEE332 pKa = 3.49KK333 pKa = 7.33WTYY336 pKa = 10.3RR337 pKa = 11.84LQNYY341 pKa = 8.36GITMTQIMSNMEE353 pKa = 3.68GSNINKK359 pKa = 9.45ILNNLEE365 pKa = 3.63KK366 pKa = 10.51QIYY369 pKa = 7.87GKK371 pKa = 10.6KK372 pKa = 8.64EE373 pKa = 3.2IKK375 pKa = 10.15YY376 pKa = 7.95YY377 pKa = 10.91ARR379 pKa = 11.84GVSMEE384 pKa = 4.21TLCC387 pKa = 5.64
MM1 pKa = 7.53LSIIKK6 pKa = 9.28NQKK9 pKa = 8.98LKK11 pKa = 11.05KK12 pKa = 10.44LIIRR16 pKa = 11.84YY17 pKa = 9.47KK18 pKa = 9.5MRR20 pKa = 11.84SLRR23 pKa = 11.84LSISKK28 pKa = 10.02SLKK31 pKa = 9.13NLKK34 pKa = 9.92EE35 pKa = 3.95NSLKK39 pKa = 10.54ARR41 pKa = 11.84KK42 pKa = 9.66AEE44 pKa = 3.85LLTRR48 pKa = 11.84LEE50 pKa = 4.16YY51 pKa = 10.54EE52 pKa = 3.6LEE54 pKa = 3.99YY55 pKa = 10.46RR56 pKa = 11.84IKK58 pKa = 10.93NGWYY62 pKa = 10.31CIFNTLTVNEE72 pKa = 4.17SSIEE76 pKa = 4.0KK77 pKa = 10.1VFEE80 pKa = 4.12KK81 pKa = 10.62GSRR84 pKa = 11.84IFSDD88 pKa = 3.72YY89 pKa = 10.69VRR91 pKa = 11.84SLDD94 pKa = 3.56RR95 pKa = 11.84GVGIAIHH102 pKa = 6.12KK103 pKa = 9.13NWRR106 pKa = 11.84QAVTKK111 pKa = 9.95RR112 pKa = 11.84KK113 pKa = 9.46EE114 pKa = 3.96GNEE117 pKa = 3.51FHH119 pKa = 6.96TYY121 pKa = 9.02FAVVEE126 pKa = 4.24RR127 pKa = 11.84GTKK130 pKa = 9.63NGRR133 pKa = 11.84LHH135 pKa = 6.1IHH137 pKa = 6.84VIHH140 pKa = 6.3MMKK143 pKa = 10.12EE144 pKa = 4.08LPNGCVDD151 pKa = 4.06PNAGRR156 pKa = 11.84AIPNRR161 pKa = 11.84RR162 pKa = 11.84EE163 pKa = 3.44VTYY166 pKa = 10.56LKK168 pKa = 10.42RR169 pKa = 11.84YY170 pKa = 6.75WKK172 pKa = 10.01YY173 pKa = 10.93GYY175 pKa = 9.36SAPIAVRR182 pKa = 11.84FNTNDD187 pKa = 3.29AFGKK191 pKa = 10.08KK192 pKa = 6.95YY193 pKa = 9.55WRR195 pKa = 11.84FPVKK199 pKa = 10.06EE200 pKa = 3.83VAKK203 pKa = 10.83NRR205 pKa = 11.84FEE207 pKa = 4.65SLEE210 pKa = 4.22CKK212 pKa = 10.28DD213 pKa = 3.49AGSIIGYY220 pKa = 8.39IGKK223 pKa = 10.04YY224 pKa = 5.93MTKK227 pKa = 10.05EE228 pKa = 3.48YY229 pKa = 10.75DD230 pKa = 3.26KK231 pKa = 10.45PLEE234 pKa = 4.18KK235 pKa = 10.35EE236 pKa = 4.05YY237 pKa = 10.0VTWRR241 pKa = 11.84TRR243 pKa = 11.84LTRR246 pKa = 11.84TLGTQIMKK254 pKa = 9.99SLCRR258 pKa = 11.84VVTKK262 pKa = 10.85NQLLLLMTMKK272 pKa = 9.38STKK275 pKa = 8.21EE276 pKa = 3.68MKK278 pKa = 10.43VLEE281 pKa = 4.76KK282 pKa = 10.39PLPLYY287 pKa = 10.45KK288 pKa = 10.43LRR290 pKa = 11.84TIATQEE296 pKa = 3.65YY297 pKa = 8.27MRR299 pKa = 11.84RR300 pKa = 11.84SLTNQPLKK308 pKa = 10.16IWEE311 pKa = 4.32SLICLTPRR319 pKa = 11.84MSLLTHH325 pKa = 6.05VKK327 pKa = 10.84SMIQEE332 pKa = 3.49KK333 pKa = 7.33WTYY336 pKa = 10.3RR337 pKa = 11.84LQNYY341 pKa = 8.36GITMTQIMSNMEE353 pKa = 3.68GSNINKK359 pKa = 9.45ILNNLEE365 pKa = 3.63KK366 pKa = 10.51QIYY369 pKa = 7.87GKK371 pKa = 10.6KK372 pKa = 8.64EE373 pKa = 3.2IKK375 pKa = 10.15YY376 pKa = 7.95YY377 pKa = 10.91ARR379 pKa = 11.84GVSMEE384 pKa = 4.21TLCC387 pKa = 5.64
Molecular weight: 45.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1824 |
59 |
428 |
202.7 |
23.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.798 ± 1.455 | 0.932 ± 0.22 |
5.099 ± 1.031 | 6.086 ± 0.704 |
3.618 ± 0.458 | 5.428 ± 0.688 |
1.7 ± 0.368 | 7.292 ± 0.514 |
8.991 ± 1.694 | 8.717 ± 0.752 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.412 ± 0.481 | 6.14 ± 0.475 |
2.961 ± 0.428 | 3.673 ± 0.626 |
4.825 ± 0.75 | 7.182 ± 0.711 |
6.579 ± 0.639 | 5.482 ± 0.656 |
1.371 ± 0.31 | 4.715 ± 0.743 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
