
Slackia equolifaciens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Slackia
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
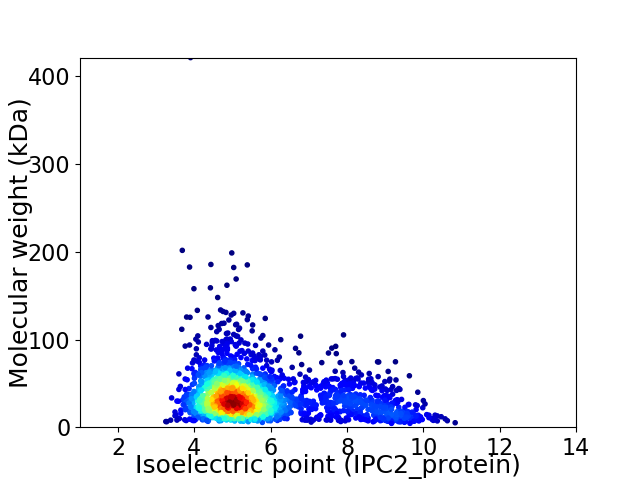
Virtual 2D-PAGE plot for 2149 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
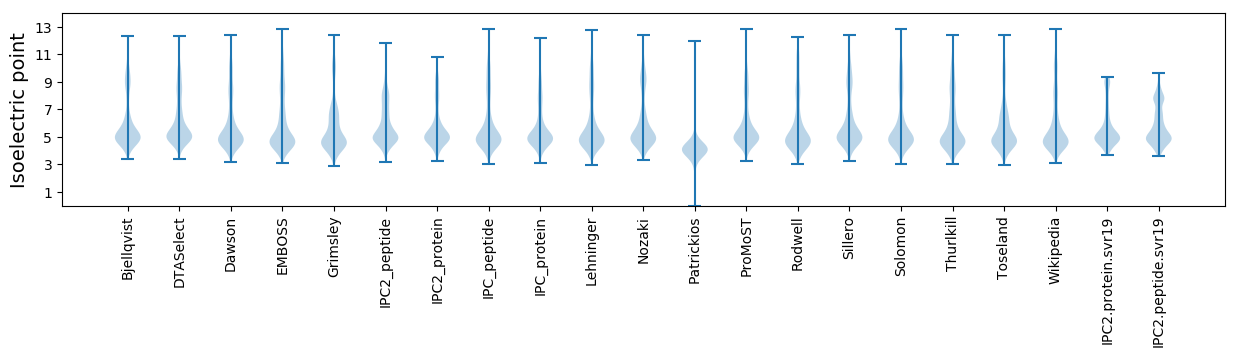
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N0AR58|A0A3N0AR58_9ACTN Protein kinase domain-containing protein OS=Slackia equolifaciens OX=498718 GN=DMP06_10985 PE=4 SV=1
MM1 pKa = 7.26VPTMLNQRR9 pKa = 11.84SASRR13 pKa = 11.84GRR15 pKa = 11.84SRR17 pKa = 11.84GRR19 pKa = 11.84VLKK22 pKa = 9.5QQPQGMKK29 pKa = 7.49RR30 pKa = 11.84TRR32 pKa = 11.84RR33 pKa = 11.84AFVALLAAVVLAFGVGSAAYY53 pKa = 9.54ANPIDD58 pKa = 4.38DD59 pKa = 3.68VVGAVTSFFTGSSNDD74 pKa = 3.35EE75 pKa = 4.02SSDD78 pKa = 3.63AGVSTQAVEE87 pKa = 4.69DD88 pKa = 3.92YY89 pKa = 9.84TVTGVSPRR97 pKa = 11.84GTTINVFDD105 pKa = 3.93YY106 pKa = 10.5WITSQDD112 pKa = 3.47DD113 pKa = 3.83PDD115 pKa = 4.25NVTQALVNDD124 pKa = 3.56QGEE127 pKa = 4.2LNEE130 pKa = 4.89RR131 pKa = 11.84YY132 pKa = 9.53KK133 pKa = 10.89QGINADD139 pKa = 3.47HH140 pKa = 6.28TLKK143 pKa = 10.83FGAGMGTSTDD153 pKa = 3.54NNEE156 pKa = 3.93DD157 pKa = 3.92LINRR161 pKa = 11.84RR162 pKa = 11.84DD163 pKa = 3.38INYY166 pKa = 6.86WTGDD170 pKa = 3.4SAPRR174 pKa = 11.84QGIVSDD180 pKa = 4.22TLTDD184 pKa = 4.78GYY186 pKa = 8.8PTLSNAGGIGSEE198 pKa = 3.99SLAYY202 pKa = 10.83LFDD205 pKa = 4.28NSTQEE210 pKa = 3.75GKK212 pKa = 9.97AAYY215 pKa = 9.02MDD217 pKa = 3.71VDD219 pKa = 3.81GLLQVDD225 pKa = 3.82DD226 pKa = 3.65QGYY229 pKa = 9.93YY230 pKa = 10.55YY231 pKa = 11.15YY232 pKa = 11.18NSQDD236 pKa = 2.93NFAQFNADD244 pKa = 3.19GSGSGNFTLYY254 pKa = 10.67NRR256 pKa = 11.84WGVEE260 pKa = 4.11AAGSSPDD267 pKa = 3.43GQFFPFNTGDD277 pKa = 3.65EE278 pKa = 4.5VFDD281 pKa = 3.92EE282 pKa = 4.43QNGQLVEE289 pKa = 4.2QQVNTSQGRR298 pKa = 11.84RR299 pKa = 11.84DD300 pKa = 4.24LISTDD305 pKa = 3.33EE306 pKa = 4.45VINHH310 pKa = 5.45YY311 pKa = 10.51FGLNMSTRR319 pKa = 11.84FIQQNGGYY327 pKa = 6.38TAPDD331 pKa = 3.71GTQGRR336 pKa = 11.84QQVTYY341 pKa = 10.33NFSGDD346 pKa = 3.73DD347 pKa = 3.96DD348 pKa = 3.79VWIYY352 pKa = 10.79IDD354 pKa = 3.94GVLVGDD360 pKa = 4.85LGGIHH365 pKa = 7.62DD366 pKa = 4.39ATSIEE371 pKa = 4.29INFATGRR378 pKa = 11.84VYY380 pKa = 10.26IYY382 pKa = 10.9DD383 pKa = 4.7DD384 pKa = 3.72GTFGDD389 pKa = 3.86NDD391 pKa = 3.44SRR393 pKa = 11.84SNNNEE398 pKa = 3.36YY399 pKa = 10.92DD400 pKa = 3.51QGEE403 pKa = 4.3TLYY406 pKa = 11.06NGSNGQTIAEE416 pKa = 4.26IMRR419 pKa = 11.84AAGVTTGLTDD429 pKa = 3.11TTFADD434 pKa = 4.06DD435 pKa = 4.15TYY437 pKa = 10.33HH438 pKa = 6.4TLDD441 pKa = 3.45FFYY444 pKa = 11.1LEE446 pKa = 4.71RR447 pKa = 11.84GNTDD451 pKa = 2.73SNMSLKK457 pKa = 10.67YY458 pKa = 10.24NLVNIPEE465 pKa = 4.16SGIVKK470 pKa = 9.48VDD472 pKa = 3.29QYY474 pKa = 11.69GDD476 pKa = 3.48EE477 pKa = 4.11LAGVQFTLSEE487 pKa = 4.2ANSNYY492 pKa = 9.54EE493 pKa = 4.29AADD496 pKa = 3.65NNNDD500 pKa = 3.35GVVTEE505 pKa = 4.09VSGRR509 pKa = 11.84TDD511 pKa = 3.18SNGEE515 pKa = 3.95MIFTYY520 pKa = 10.29EE521 pKa = 4.06NNSGQEE527 pKa = 3.81MPITLEE533 pKa = 3.86QMGEE537 pKa = 3.86RR538 pKa = 11.84SAYY541 pKa = 8.97WILEE545 pKa = 3.96EE546 pKa = 5.37AEE548 pKa = 4.19GNEE551 pKa = 4.31TIGYY555 pKa = 7.52RR556 pKa = 11.84NPGTVEE562 pKa = 3.97LRR564 pKa = 11.84FSTINGDD571 pKa = 3.57GTGDD575 pKa = 3.65GVLLSSNQWEE585 pKa = 4.3TGAYY589 pKa = 8.82SQAHH593 pKa = 4.65VTATAPTNVTAVDD606 pKa = 3.23NGGTYY611 pKa = 10.31EE612 pKa = 4.23SAGSLMFAVVAQIDD626 pKa = 3.92EE627 pKa = 4.25QGNYY631 pKa = 9.59HH632 pKa = 6.98AVTGDD637 pKa = 3.75AFAGWSVADD646 pKa = 3.86ATIDD650 pKa = 3.85DD651 pKa = 4.5ASTSKK656 pKa = 10.95DD657 pKa = 3.63AIVTAAKK664 pKa = 10.38ASGYY668 pKa = 10.23QFLLGSGGAYY678 pKa = 9.3QVTIDD683 pKa = 5.15DD684 pKa = 4.67LPGDD688 pKa = 3.92INTYY692 pKa = 10.35EE693 pKa = 3.96YY694 pKa = 10.23MIEE697 pKa = 4.03QAGGDD702 pKa = 3.69TSTAQYY708 pKa = 9.77AVKK711 pKa = 10.35YY712 pKa = 8.38YY713 pKa = 10.04WSDD716 pKa = 2.99ATSIDD721 pKa = 4.05DD722 pKa = 4.64LSSNSQIVEE731 pKa = 3.83IDD733 pKa = 3.67PNTGDD738 pKa = 3.08GFEE741 pKa = 5.26RR742 pKa = 11.84IFSVTLSIPNIKK754 pKa = 10.44NEE756 pKa = 3.77LSLVKK761 pKa = 9.89TDD763 pKa = 3.83EE764 pKa = 4.42TTGDD768 pKa = 4.18PIAGVVFNMYY778 pKa = 9.83EE779 pKa = 4.15DD780 pKa = 4.0KK781 pKa = 11.53NGDD784 pKa = 3.04GTYY787 pKa = 11.15NNGEE791 pKa = 4.18RR792 pKa = 11.84CCFTGTTGTDD802 pKa = 2.93GGLEE806 pKa = 3.67ITSNINNGEE815 pKa = 3.95QVLAKK820 pKa = 10.42GEE822 pKa = 4.21YY823 pKa = 9.8VLVEE827 pKa = 4.37TVPSGYY833 pKa = 10.15VDD835 pKa = 3.64EE836 pKa = 4.39QTSIRR841 pKa = 11.84VIVDD845 pKa = 3.17DD846 pKa = 5.68DD847 pKa = 4.35GVHH850 pKa = 6.51VDD852 pKa = 5.5AGDD855 pKa = 3.92ANDD858 pKa = 3.57NVTVEE863 pKa = 4.18TGIGSLVYY871 pKa = 10.77SMRR874 pKa = 11.84GFAAGDD880 pKa = 3.82DD881 pKa = 4.13VDD883 pKa = 4.38STLHH887 pKa = 6.81DD888 pKa = 3.73VVAQAQTADD897 pKa = 3.76SYY899 pKa = 11.57EE900 pKa = 4.36GASTAWDD907 pKa = 3.31NADD910 pKa = 3.73AVTHH914 pKa = 5.31YY915 pKa = 10.73QYY917 pKa = 11.62DD918 pKa = 3.88DD919 pKa = 3.34QTDD922 pKa = 3.56GLNYY926 pKa = 9.53IRR928 pKa = 11.84NDD930 pKa = 3.13NVTDD934 pKa = 3.63NGDD937 pKa = 3.23ATFIADD943 pKa = 4.13AGWSRR948 pKa = 11.84LDD950 pKa = 3.82LRR952 pKa = 11.84QCTAHH957 pKa = 6.94AGEE960 pKa = 5.15DD961 pKa = 3.87PIEE964 pKa = 4.31NKK966 pKa = 9.99QNLGDD971 pKa = 3.61QSLNALFTGDD981 pKa = 3.44VTIHH985 pKa = 5.19VTNSQIPTTSSLTISKK1001 pKa = 8.63TVTGTGAPSDD1011 pKa = 3.7VTFDD1015 pKa = 3.79FSFSLTDD1022 pKa = 3.24AGGNSVTGTFSATTHH1037 pKa = 6.49RR1038 pKa = 11.84ADD1040 pKa = 3.78GTEE1043 pKa = 3.9STADD1047 pKa = 3.19MVIYY1051 pKa = 10.89AEE1053 pKa = 4.53ADD1055 pKa = 3.7CNQLTLRR1062 pKa = 11.84NGEE1065 pKa = 4.26SVTIDD1070 pKa = 3.66GLPISANYY1078 pKa = 8.31TVTEE1082 pKa = 4.08EE1083 pKa = 4.0AKK1085 pKa = 10.48GYY1087 pKa = 7.06YY1088 pKa = 8.6TPSVQQSSGTLNEE1101 pKa = 5.1DD1102 pKa = 3.53GSFTATGTIDD1112 pKa = 3.83NDD1114 pKa = 3.61GATAAFSNDD1123 pKa = 2.67FWGGSVDD1130 pKa = 3.59YY1131 pKa = 10.6DD1132 pKa = 3.63ASIDD1136 pKa = 3.79LTVSKK1141 pKa = 11.13VMTGRR1146 pKa = 11.84DD1147 pKa = 3.17MTAGEE1152 pKa = 4.31FAVVVDD1158 pKa = 5.2PINDD1162 pKa = 3.2EE1163 pKa = 4.19SAALLGLANASEE1175 pKa = 4.36NKK1177 pKa = 9.86VIDD1180 pKa = 3.67MPAANDD1186 pKa = 3.99GVEE1189 pKa = 4.06VSVDD1193 pKa = 3.11ILNGLNDD1200 pKa = 3.42IAFTAEE1206 pKa = 4.12DD1207 pKa = 3.69ADD1209 pKa = 3.78KK1210 pKa = 10.47TFTYY1214 pKa = 9.41TVYY1217 pKa = 10.42EE1218 pKa = 4.16QQSDD1222 pKa = 3.87QNPSNGVTYY1231 pKa = 8.54DD1232 pKa = 3.3TARR1235 pKa = 11.84YY1236 pKa = 9.24SLTIEE1241 pKa = 4.13VSVDD1245 pKa = 2.72ADD1247 pKa = 3.53GVVTVHH1253 pKa = 5.92TAVSNEE1259 pKa = 3.77TGTLRR1264 pKa = 11.84EE1265 pKa = 4.12MTTTPNSGGTMPVAVFNNSYY1285 pKa = 10.7NATGEE1290 pKa = 4.08LGGNGSASIKK1300 pKa = 8.36ATKK1303 pKa = 10.29VLEE1306 pKa = 4.22NDD1308 pKa = 3.25TLAAGDD1314 pKa = 3.65FTFNVAAVNSNGTDD1328 pKa = 3.22EE1329 pKa = 4.91RR1330 pKa = 11.84VEE1332 pKa = 3.97ATTTNGTNGAASAADD1347 pKa = 3.23IVFPAIQYY1355 pKa = 10.85DD1356 pKa = 4.18SDD1358 pKa = 4.14SLAQDD1363 pKa = 3.27VADD1366 pKa = 5.08GYY1368 pKa = 11.86ARR1370 pKa = 11.84ARR1372 pKa = 11.84VNDD1375 pKa = 3.81NGTTTYY1381 pKa = 9.59TYY1383 pKa = 10.64LYY1385 pKa = 9.8RR1386 pKa = 11.84VYY1388 pKa = 10.46EE1389 pKa = 4.18DD1390 pKa = 3.23TSGFDD1395 pKa = 3.7GLGLTTSQSSYY1406 pKa = 11.23NVTVWVTDD1414 pKa = 3.97DD1415 pKa = 3.9GDD1417 pKa = 3.74GSLEE1421 pKa = 4.16VEE1423 pKa = 4.14VGYY1426 pKa = 8.42PTGSEE1431 pKa = 3.73NGITFTNVYY1440 pKa = 9.25GASANATLNVGGTKK1454 pKa = 9.23IYY1456 pKa = 7.57EE1457 pKa = 4.28TNGLKK1462 pKa = 10.09NAPSIEE1468 pKa = 3.93GKK1470 pKa = 8.55YY1471 pKa = 8.48TFTLTGVDD1479 pKa = 3.55EE1480 pKa = 5.5DD1481 pKa = 4.81GNAAPLPGGATGSVTAQNQANGTVDD1506 pKa = 4.64FGTITYY1512 pKa = 10.06DD1513 pKa = 3.19IADD1516 pKa = 4.3LGGADD1521 pKa = 4.14SKK1523 pKa = 10.28TFTYY1527 pKa = 10.24TVTEE1531 pKa = 4.05SGGVDD1536 pKa = 3.25GVTNDD1541 pKa = 3.67GAASTGKK1548 pKa = 8.16TFTVTLTDD1556 pKa = 3.44NGKK1559 pKa = 8.4GTISVDD1565 pKa = 3.22SDD1567 pKa = 3.45ALQAAGSQFSFTNEE1581 pKa = 3.76YY1582 pKa = 10.85SVDD1585 pKa = 3.64PTDD1588 pKa = 4.04PTDD1591 pKa = 3.79PTDD1594 pKa = 4.24PGTDD1598 pKa = 3.3PSDD1601 pKa = 3.53PTTAALSISKK1611 pKa = 10.04VLTGRR1616 pKa = 11.84AMNEE1620 pKa = 4.1GEE1622 pKa = 4.85FSFTMAAAGDD1632 pKa = 3.88YY1633 pKa = 10.62GNAVSPTSITATNMVNEE1650 pKa = 5.15DD1651 pKa = 3.32GTGSVVFGDD1660 pKa = 3.45GFVFTEE1666 pKa = 4.03PGTYY1670 pKa = 10.17HH1671 pKa = 5.47FTISEE1676 pKa = 4.24DD1677 pKa = 3.34VGTLGGVSYY1686 pKa = 10.99DD1687 pKa = 3.39GSDD1690 pKa = 3.55YY1691 pKa = 10.86TATAVVTDD1699 pKa = 4.91DD1700 pKa = 3.6GAGNLSVAWTVADD1713 pKa = 3.66AEE1715 pKa = 4.59GNAVSEE1721 pKa = 4.23ITFEE1725 pKa = 4.03NTYY1728 pKa = 7.14TTSATTPVEE1737 pKa = 4.0LKK1739 pKa = 10.57VSKK1742 pKa = 10.05EE1743 pKa = 4.07LQDD1746 pKa = 3.32GTLADD1751 pKa = 3.88GQFTFEE1757 pKa = 4.39LTGSDD1762 pKa = 4.07GAPMPEE1768 pKa = 4.2STTVTNNAQGVVSFGSITFDD1788 pKa = 3.34AVGEE1792 pKa = 3.95YY1793 pKa = 10.02DD1794 pKa = 3.75YY1795 pKa = 11.56TVTEE1799 pKa = 4.4VNDD1802 pKa = 4.27GQDD1805 pKa = 3.42GITYY1809 pKa = 10.53DD1810 pKa = 3.81EE1811 pKa = 4.89DD1812 pKa = 3.41ATRR1815 pKa = 11.84TIHH1818 pKa = 7.16VSVTDD1823 pKa = 3.88DD1824 pKa = 4.2GEE1826 pKa = 4.3GHH1828 pKa = 6.93LVAEE1832 pKa = 4.49VTYY1835 pKa = 10.7GSDD1838 pKa = 2.98GSHH1841 pKa = 6.04FVNVNTTGGNNDD1853 pKa = 3.74GGDD1856 pKa = 3.63NGDD1859 pKa = 4.23GGNGGDD1865 pKa = 4.16NGGTGDD1871 pKa = 4.32AGNGSGNGSSFAQTNDD1887 pKa = 2.93ATPWPLAMALALVAAGAIIAAVFGLRR1913 pKa = 11.84RR1914 pKa = 11.84AQRR1917 pKa = 3.33
MM1 pKa = 7.26VPTMLNQRR9 pKa = 11.84SASRR13 pKa = 11.84GRR15 pKa = 11.84SRR17 pKa = 11.84GRR19 pKa = 11.84VLKK22 pKa = 9.5QQPQGMKK29 pKa = 7.49RR30 pKa = 11.84TRR32 pKa = 11.84RR33 pKa = 11.84AFVALLAAVVLAFGVGSAAYY53 pKa = 9.54ANPIDD58 pKa = 4.38DD59 pKa = 3.68VVGAVTSFFTGSSNDD74 pKa = 3.35EE75 pKa = 4.02SSDD78 pKa = 3.63AGVSTQAVEE87 pKa = 4.69DD88 pKa = 3.92YY89 pKa = 9.84TVTGVSPRR97 pKa = 11.84GTTINVFDD105 pKa = 3.93YY106 pKa = 10.5WITSQDD112 pKa = 3.47DD113 pKa = 3.83PDD115 pKa = 4.25NVTQALVNDD124 pKa = 3.56QGEE127 pKa = 4.2LNEE130 pKa = 4.89RR131 pKa = 11.84YY132 pKa = 9.53KK133 pKa = 10.89QGINADD139 pKa = 3.47HH140 pKa = 6.28TLKK143 pKa = 10.83FGAGMGTSTDD153 pKa = 3.54NNEE156 pKa = 3.93DD157 pKa = 3.92LINRR161 pKa = 11.84RR162 pKa = 11.84DD163 pKa = 3.38INYY166 pKa = 6.86WTGDD170 pKa = 3.4SAPRR174 pKa = 11.84QGIVSDD180 pKa = 4.22TLTDD184 pKa = 4.78GYY186 pKa = 8.8PTLSNAGGIGSEE198 pKa = 3.99SLAYY202 pKa = 10.83LFDD205 pKa = 4.28NSTQEE210 pKa = 3.75GKK212 pKa = 9.97AAYY215 pKa = 9.02MDD217 pKa = 3.71VDD219 pKa = 3.81GLLQVDD225 pKa = 3.82DD226 pKa = 3.65QGYY229 pKa = 9.93YY230 pKa = 10.55YY231 pKa = 11.15YY232 pKa = 11.18NSQDD236 pKa = 2.93NFAQFNADD244 pKa = 3.19GSGSGNFTLYY254 pKa = 10.67NRR256 pKa = 11.84WGVEE260 pKa = 4.11AAGSSPDD267 pKa = 3.43GQFFPFNTGDD277 pKa = 3.65EE278 pKa = 4.5VFDD281 pKa = 3.92EE282 pKa = 4.43QNGQLVEE289 pKa = 4.2QQVNTSQGRR298 pKa = 11.84RR299 pKa = 11.84DD300 pKa = 4.24LISTDD305 pKa = 3.33EE306 pKa = 4.45VINHH310 pKa = 5.45YY311 pKa = 10.51FGLNMSTRR319 pKa = 11.84FIQQNGGYY327 pKa = 6.38TAPDD331 pKa = 3.71GTQGRR336 pKa = 11.84QQVTYY341 pKa = 10.33NFSGDD346 pKa = 3.73DD347 pKa = 3.96DD348 pKa = 3.79VWIYY352 pKa = 10.79IDD354 pKa = 3.94GVLVGDD360 pKa = 4.85LGGIHH365 pKa = 7.62DD366 pKa = 4.39ATSIEE371 pKa = 4.29INFATGRR378 pKa = 11.84VYY380 pKa = 10.26IYY382 pKa = 10.9DD383 pKa = 4.7DD384 pKa = 3.72GTFGDD389 pKa = 3.86NDD391 pKa = 3.44SRR393 pKa = 11.84SNNNEE398 pKa = 3.36YY399 pKa = 10.92DD400 pKa = 3.51QGEE403 pKa = 4.3TLYY406 pKa = 11.06NGSNGQTIAEE416 pKa = 4.26IMRR419 pKa = 11.84AAGVTTGLTDD429 pKa = 3.11TTFADD434 pKa = 4.06DD435 pKa = 4.15TYY437 pKa = 10.33HH438 pKa = 6.4TLDD441 pKa = 3.45FFYY444 pKa = 11.1LEE446 pKa = 4.71RR447 pKa = 11.84GNTDD451 pKa = 2.73SNMSLKK457 pKa = 10.67YY458 pKa = 10.24NLVNIPEE465 pKa = 4.16SGIVKK470 pKa = 9.48VDD472 pKa = 3.29QYY474 pKa = 11.69GDD476 pKa = 3.48EE477 pKa = 4.11LAGVQFTLSEE487 pKa = 4.2ANSNYY492 pKa = 9.54EE493 pKa = 4.29AADD496 pKa = 3.65NNNDD500 pKa = 3.35GVVTEE505 pKa = 4.09VSGRR509 pKa = 11.84TDD511 pKa = 3.18SNGEE515 pKa = 3.95MIFTYY520 pKa = 10.29EE521 pKa = 4.06NNSGQEE527 pKa = 3.81MPITLEE533 pKa = 3.86QMGEE537 pKa = 3.86RR538 pKa = 11.84SAYY541 pKa = 8.97WILEE545 pKa = 3.96EE546 pKa = 5.37AEE548 pKa = 4.19GNEE551 pKa = 4.31TIGYY555 pKa = 7.52RR556 pKa = 11.84NPGTVEE562 pKa = 3.97LRR564 pKa = 11.84FSTINGDD571 pKa = 3.57GTGDD575 pKa = 3.65GVLLSSNQWEE585 pKa = 4.3TGAYY589 pKa = 8.82SQAHH593 pKa = 4.65VTATAPTNVTAVDD606 pKa = 3.23NGGTYY611 pKa = 10.31EE612 pKa = 4.23SAGSLMFAVVAQIDD626 pKa = 3.92EE627 pKa = 4.25QGNYY631 pKa = 9.59HH632 pKa = 6.98AVTGDD637 pKa = 3.75AFAGWSVADD646 pKa = 3.86ATIDD650 pKa = 3.85DD651 pKa = 4.5ASTSKK656 pKa = 10.95DD657 pKa = 3.63AIVTAAKK664 pKa = 10.38ASGYY668 pKa = 10.23QFLLGSGGAYY678 pKa = 9.3QVTIDD683 pKa = 5.15DD684 pKa = 4.67LPGDD688 pKa = 3.92INTYY692 pKa = 10.35EE693 pKa = 3.96YY694 pKa = 10.23MIEE697 pKa = 4.03QAGGDD702 pKa = 3.69TSTAQYY708 pKa = 9.77AVKK711 pKa = 10.35YY712 pKa = 8.38YY713 pKa = 10.04WSDD716 pKa = 2.99ATSIDD721 pKa = 4.05DD722 pKa = 4.64LSSNSQIVEE731 pKa = 3.83IDD733 pKa = 3.67PNTGDD738 pKa = 3.08GFEE741 pKa = 5.26RR742 pKa = 11.84IFSVTLSIPNIKK754 pKa = 10.44NEE756 pKa = 3.77LSLVKK761 pKa = 9.89TDD763 pKa = 3.83EE764 pKa = 4.42TTGDD768 pKa = 4.18PIAGVVFNMYY778 pKa = 9.83EE779 pKa = 4.15DD780 pKa = 4.0KK781 pKa = 11.53NGDD784 pKa = 3.04GTYY787 pKa = 11.15NNGEE791 pKa = 4.18RR792 pKa = 11.84CCFTGTTGTDD802 pKa = 2.93GGLEE806 pKa = 3.67ITSNINNGEE815 pKa = 3.95QVLAKK820 pKa = 10.42GEE822 pKa = 4.21YY823 pKa = 9.8VLVEE827 pKa = 4.37TVPSGYY833 pKa = 10.15VDD835 pKa = 3.64EE836 pKa = 4.39QTSIRR841 pKa = 11.84VIVDD845 pKa = 3.17DD846 pKa = 5.68DD847 pKa = 4.35GVHH850 pKa = 6.51VDD852 pKa = 5.5AGDD855 pKa = 3.92ANDD858 pKa = 3.57NVTVEE863 pKa = 4.18TGIGSLVYY871 pKa = 10.77SMRR874 pKa = 11.84GFAAGDD880 pKa = 3.82DD881 pKa = 4.13VDD883 pKa = 4.38STLHH887 pKa = 6.81DD888 pKa = 3.73VVAQAQTADD897 pKa = 3.76SYY899 pKa = 11.57EE900 pKa = 4.36GASTAWDD907 pKa = 3.31NADD910 pKa = 3.73AVTHH914 pKa = 5.31YY915 pKa = 10.73QYY917 pKa = 11.62DD918 pKa = 3.88DD919 pKa = 3.34QTDD922 pKa = 3.56GLNYY926 pKa = 9.53IRR928 pKa = 11.84NDD930 pKa = 3.13NVTDD934 pKa = 3.63NGDD937 pKa = 3.23ATFIADD943 pKa = 4.13AGWSRR948 pKa = 11.84LDD950 pKa = 3.82LRR952 pKa = 11.84QCTAHH957 pKa = 6.94AGEE960 pKa = 5.15DD961 pKa = 3.87PIEE964 pKa = 4.31NKK966 pKa = 9.99QNLGDD971 pKa = 3.61QSLNALFTGDD981 pKa = 3.44VTIHH985 pKa = 5.19VTNSQIPTTSSLTISKK1001 pKa = 8.63TVTGTGAPSDD1011 pKa = 3.7VTFDD1015 pKa = 3.79FSFSLTDD1022 pKa = 3.24AGGNSVTGTFSATTHH1037 pKa = 6.49RR1038 pKa = 11.84ADD1040 pKa = 3.78GTEE1043 pKa = 3.9STADD1047 pKa = 3.19MVIYY1051 pKa = 10.89AEE1053 pKa = 4.53ADD1055 pKa = 3.7CNQLTLRR1062 pKa = 11.84NGEE1065 pKa = 4.26SVTIDD1070 pKa = 3.66GLPISANYY1078 pKa = 8.31TVTEE1082 pKa = 4.08EE1083 pKa = 4.0AKK1085 pKa = 10.48GYY1087 pKa = 7.06YY1088 pKa = 8.6TPSVQQSSGTLNEE1101 pKa = 5.1DD1102 pKa = 3.53GSFTATGTIDD1112 pKa = 3.83NDD1114 pKa = 3.61GATAAFSNDD1123 pKa = 2.67FWGGSVDD1130 pKa = 3.59YY1131 pKa = 10.6DD1132 pKa = 3.63ASIDD1136 pKa = 3.79LTVSKK1141 pKa = 11.13VMTGRR1146 pKa = 11.84DD1147 pKa = 3.17MTAGEE1152 pKa = 4.31FAVVVDD1158 pKa = 5.2PINDD1162 pKa = 3.2EE1163 pKa = 4.19SAALLGLANASEE1175 pKa = 4.36NKK1177 pKa = 9.86VIDD1180 pKa = 3.67MPAANDD1186 pKa = 3.99GVEE1189 pKa = 4.06VSVDD1193 pKa = 3.11ILNGLNDD1200 pKa = 3.42IAFTAEE1206 pKa = 4.12DD1207 pKa = 3.69ADD1209 pKa = 3.78KK1210 pKa = 10.47TFTYY1214 pKa = 9.41TVYY1217 pKa = 10.42EE1218 pKa = 4.16QQSDD1222 pKa = 3.87QNPSNGVTYY1231 pKa = 8.54DD1232 pKa = 3.3TARR1235 pKa = 11.84YY1236 pKa = 9.24SLTIEE1241 pKa = 4.13VSVDD1245 pKa = 2.72ADD1247 pKa = 3.53GVVTVHH1253 pKa = 5.92TAVSNEE1259 pKa = 3.77TGTLRR1264 pKa = 11.84EE1265 pKa = 4.12MTTTPNSGGTMPVAVFNNSYY1285 pKa = 10.7NATGEE1290 pKa = 4.08LGGNGSASIKK1300 pKa = 8.36ATKK1303 pKa = 10.29VLEE1306 pKa = 4.22NDD1308 pKa = 3.25TLAAGDD1314 pKa = 3.65FTFNVAAVNSNGTDD1328 pKa = 3.22EE1329 pKa = 4.91RR1330 pKa = 11.84VEE1332 pKa = 3.97ATTTNGTNGAASAADD1347 pKa = 3.23IVFPAIQYY1355 pKa = 10.85DD1356 pKa = 4.18SDD1358 pKa = 4.14SLAQDD1363 pKa = 3.27VADD1366 pKa = 5.08GYY1368 pKa = 11.86ARR1370 pKa = 11.84ARR1372 pKa = 11.84VNDD1375 pKa = 3.81NGTTTYY1381 pKa = 9.59TYY1383 pKa = 10.64LYY1385 pKa = 9.8RR1386 pKa = 11.84VYY1388 pKa = 10.46EE1389 pKa = 4.18DD1390 pKa = 3.23TSGFDD1395 pKa = 3.7GLGLTTSQSSYY1406 pKa = 11.23NVTVWVTDD1414 pKa = 3.97DD1415 pKa = 3.9GDD1417 pKa = 3.74GSLEE1421 pKa = 4.16VEE1423 pKa = 4.14VGYY1426 pKa = 8.42PTGSEE1431 pKa = 3.73NGITFTNVYY1440 pKa = 9.25GASANATLNVGGTKK1454 pKa = 9.23IYY1456 pKa = 7.57EE1457 pKa = 4.28TNGLKK1462 pKa = 10.09NAPSIEE1468 pKa = 3.93GKK1470 pKa = 8.55YY1471 pKa = 8.48TFTLTGVDD1479 pKa = 3.55EE1480 pKa = 5.5DD1481 pKa = 4.81GNAAPLPGGATGSVTAQNQANGTVDD1506 pKa = 4.64FGTITYY1512 pKa = 10.06DD1513 pKa = 3.19IADD1516 pKa = 4.3LGGADD1521 pKa = 4.14SKK1523 pKa = 10.28TFTYY1527 pKa = 10.24TVTEE1531 pKa = 4.05SGGVDD1536 pKa = 3.25GVTNDD1541 pKa = 3.67GAASTGKK1548 pKa = 8.16TFTVTLTDD1556 pKa = 3.44NGKK1559 pKa = 8.4GTISVDD1565 pKa = 3.22SDD1567 pKa = 3.45ALQAAGSQFSFTNEE1581 pKa = 3.76YY1582 pKa = 10.85SVDD1585 pKa = 3.64PTDD1588 pKa = 4.04PTDD1591 pKa = 3.79PTDD1594 pKa = 4.24PGTDD1598 pKa = 3.3PSDD1601 pKa = 3.53PTTAALSISKK1611 pKa = 10.04VLTGRR1616 pKa = 11.84AMNEE1620 pKa = 4.1GEE1622 pKa = 4.85FSFTMAAAGDD1632 pKa = 3.88YY1633 pKa = 10.62GNAVSPTSITATNMVNEE1650 pKa = 5.15DD1651 pKa = 3.32GTGSVVFGDD1660 pKa = 3.45GFVFTEE1666 pKa = 4.03PGTYY1670 pKa = 10.17HH1671 pKa = 5.47FTISEE1676 pKa = 4.24DD1677 pKa = 3.34VGTLGGVSYY1686 pKa = 10.99DD1687 pKa = 3.39GSDD1690 pKa = 3.55YY1691 pKa = 10.86TATAVVTDD1699 pKa = 4.91DD1700 pKa = 3.6GAGNLSVAWTVADD1713 pKa = 3.66AEE1715 pKa = 4.59GNAVSEE1721 pKa = 4.23ITFEE1725 pKa = 4.03NTYY1728 pKa = 7.14TTSATTPVEE1737 pKa = 4.0LKK1739 pKa = 10.57VSKK1742 pKa = 10.05EE1743 pKa = 4.07LQDD1746 pKa = 3.32GTLADD1751 pKa = 3.88GQFTFEE1757 pKa = 4.39LTGSDD1762 pKa = 4.07GAPMPEE1768 pKa = 4.2STTVTNNAQGVVSFGSITFDD1788 pKa = 3.34AVGEE1792 pKa = 3.95YY1793 pKa = 10.02DD1794 pKa = 3.75YY1795 pKa = 11.56TVTEE1799 pKa = 4.4VNDD1802 pKa = 4.27GQDD1805 pKa = 3.42GITYY1809 pKa = 10.53DD1810 pKa = 3.81EE1811 pKa = 4.89DD1812 pKa = 3.41ATRR1815 pKa = 11.84TIHH1818 pKa = 7.16VSVTDD1823 pKa = 3.88DD1824 pKa = 4.2GEE1826 pKa = 4.3GHH1828 pKa = 6.93LVAEE1832 pKa = 4.49VTYY1835 pKa = 10.7GSDD1838 pKa = 2.98GSHH1841 pKa = 6.04FVNVNTTGGNNDD1853 pKa = 3.74GGDD1856 pKa = 3.63NGDD1859 pKa = 4.23GGNGGDD1865 pKa = 4.16NGGTGDD1871 pKa = 4.32AGNGSGNGSSFAQTNDD1887 pKa = 2.93ATPWPLAMALALVAAGAIIAAVFGLRR1913 pKa = 11.84RR1914 pKa = 11.84AQRR1917 pKa = 3.33
Molecular weight: 201.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N0AUU4|A0A3N0AUU4_9ACTN LysR family transcriptional regulator OS=Slackia equolifaciens OX=498718 GN=DMP06_09035 PE=3 SV=1
MM1 pKa = 7.39AAAPRR6 pKa = 11.84YY7 pKa = 6.09VTCLFGSMKK16 pKa = 10.22SGKK19 pKa = 8.17FTQRR23 pKa = 11.84STEE26 pKa = 3.7AKK28 pKa = 8.83AARR31 pKa = 11.84GTFVRR36 pKa = 11.84WCAEE40 pKa = 3.51NNVRR44 pKa = 11.84HH45 pKa = 5.74VSDD48 pKa = 4.31FQHH51 pKa = 6.7FDD53 pKa = 2.75IGGYY57 pKa = 8.65QYY59 pKa = 11.66NEE61 pKa = 4.1ALSSEE66 pKa = 4.16DD67 pKa = 3.24TFVFTHH73 pKa = 5.6QGRR76 pKa = 11.84PLRR79 pKa = 11.84CPMGAAIVRR88 pKa = 11.84LGSVSGAAQEE98 pKa = 4.02ASPRR102 pKa = 11.84KK103 pKa = 9.55ARR105 pKa = 11.84SGDD108 pKa = 3.16SRR110 pKa = 11.84RR111 pKa = 11.84LRR113 pKa = 11.84RR114 pKa = 11.84PII116 pKa = 3.62
MM1 pKa = 7.39AAAPRR6 pKa = 11.84YY7 pKa = 6.09VTCLFGSMKK16 pKa = 10.22SGKK19 pKa = 8.17FTQRR23 pKa = 11.84STEE26 pKa = 3.7AKK28 pKa = 8.83AARR31 pKa = 11.84GTFVRR36 pKa = 11.84WCAEE40 pKa = 3.51NNVRR44 pKa = 11.84HH45 pKa = 5.74VSDD48 pKa = 4.31FQHH51 pKa = 6.7FDD53 pKa = 2.75IGGYY57 pKa = 8.65QYY59 pKa = 11.66NEE61 pKa = 4.1ALSSEE66 pKa = 4.16DD67 pKa = 3.24TFVFTHH73 pKa = 5.6QGRR76 pKa = 11.84PLRR79 pKa = 11.84CPMGAAIVRR88 pKa = 11.84LGSVSGAAQEE98 pKa = 4.02ASPRR102 pKa = 11.84KK103 pKa = 9.55ARR105 pKa = 11.84SGDD108 pKa = 3.16SRR110 pKa = 11.84RR111 pKa = 11.84LRR113 pKa = 11.84RR114 pKa = 11.84PII116 pKa = 3.62
Molecular weight: 12.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
742730 |
36 |
3987 |
345.6 |
37.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.816 ± 0.075 | 1.667 ± 0.028 |
6.081 ± 0.038 | 6.757 ± 0.052 |
3.847 ± 0.032 | 8.316 ± 0.058 |
1.861 ± 0.023 | 5.253 ± 0.042 |
3.785 ± 0.041 | 8.784 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.821 ± 0.025 | 2.934 ± 0.038 |
4.253 ± 0.037 | 2.981 ± 0.03 |
5.849 ± 0.061 | 6.104 ± 0.049 |
5.099 ± 0.064 | 7.865 ± 0.046 |
1.055 ± 0.019 | 2.871 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
