
Sulfitobacter sp. JBTF-M27
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Sulfitobacter
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
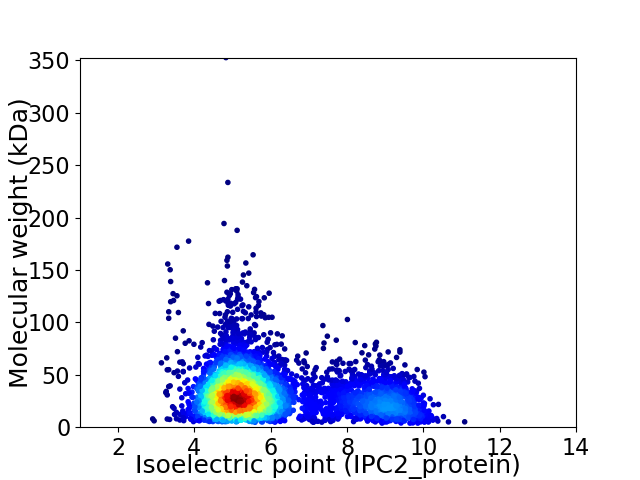
Virtual 2D-PAGE plot for 4488 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
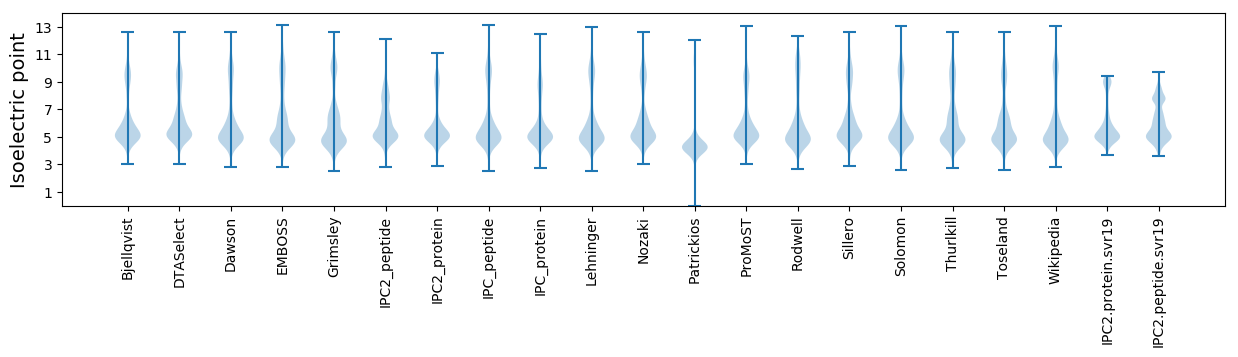
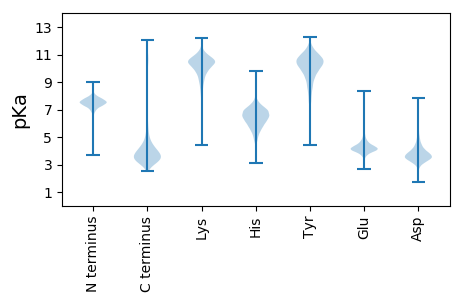
Protein with the lowest isoelectric point:
>tr|A0A6P0C9J0|A0A6P0C9J0_9RHOB Peptidoglycan DD-metalloendopeptidase family protein OS=Sulfitobacter sp. JBTF-M27 OX=2698830 GN=GV827_08940 PE=4 SV=1
MM1 pKa = 7.22FPEE4 pKa = 5.11DD5 pKa = 4.13SPLLAALASYY15 pKa = 10.94GSDD18 pKa = 2.69EE19 pKa = 4.37RR20 pKa = 11.84LYY22 pKa = 10.48TSTVTFRR29 pKa = 11.84FMPAGSEE36 pKa = 4.16IDD38 pKa = 4.81DD39 pKa = 4.6GDD41 pKa = 4.39PNTPNEE47 pKa = 4.14VSVGWSEE54 pKa = 5.03PNMQYY59 pKa = 10.77VRR61 pKa = 11.84GTFDD65 pKa = 3.81YY66 pKa = 10.42ISSVVDD72 pKa = 3.16LTFAEE77 pKa = 4.85TTSSDD82 pKa = 3.16ATISFYY88 pKa = 11.08EE89 pKa = 4.06VAEE92 pKa = 4.39FSDD95 pKa = 3.81DD96 pKa = 3.21TAGYY100 pKa = 8.23VTFISEE106 pKa = 4.81GISIAVIGSQFTGPVAFDD124 pKa = 3.46NDD126 pKa = 3.8YY127 pKa = 8.61TTIIHH132 pKa = 6.87EE133 pKa = 4.42IGHH136 pKa = 7.07AIGLSHH142 pKa = 6.99PHH144 pKa = 7.3DD145 pKa = 4.97GPAVLPGVTSQDD157 pKa = 3.44DD158 pKa = 3.8KK159 pKa = 12.09GDD161 pKa = 4.08LNLNTEE167 pKa = 4.33FTTRR171 pKa = 11.84MSYY174 pKa = 10.24IPGEE178 pKa = 4.41SPSNPGVDD186 pKa = 2.54IWGEE190 pKa = 4.0ANTFGAIDD198 pKa = 3.54IAALQLLYY206 pKa = 10.74GANTQTGLGDD216 pKa = 3.6TVYY219 pKa = 11.33GDD221 pKa = 3.43TRR223 pKa = 11.84DD224 pKa = 5.3LVTIWDD230 pKa = 3.53NGGFDD235 pKa = 5.18RR236 pKa = 11.84IDD238 pKa = 3.5FSSATEE244 pKa = 3.93DD245 pKa = 3.43AVIDD249 pKa = 3.85LRR251 pKa = 11.84AATLQFEE258 pKa = 4.75VGGGGYY264 pKa = 10.48LSYY267 pKa = 10.64ISSRR271 pKa = 11.84NGTVADD277 pKa = 3.61GGYY280 pKa = 10.23AIAYY284 pKa = 8.49GVEE287 pKa = 3.98IEE289 pKa = 4.23NAMGGAGADD298 pKa = 3.27MLMGNALANTFVGGGGNDD316 pKa = 3.56VIDD319 pKa = 4.65GGDD322 pKa = 3.56GRR324 pKa = 11.84DD325 pKa = 3.19TGAYY329 pKa = 9.71SGNQSSYY336 pKa = 10.58TLTLSAQTTTLADD349 pKa = 3.52RR350 pKa = 11.84RR351 pKa = 11.84GTDD354 pKa = 3.32GTDD357 pKa = 2.86TLVNMEE363 pKa = 5.26LLTFGDD369 pKa = 3.71AAVAPFDD376 pKa = 3.68LTVFAGTTGLSEE388 pKa = 4.14TQMEE392 pKa = 4.67SFIEE396 pKa = 4.25LYY398 pKa = 9.75IAYY401 pKa = 9.93FNRR404 pKa = 11.84APDD407 pKa = 3.65AVGLNFWGTAFASGTTLEE425 pKa = 4.12QMATLFTDD433 pKa = 3.29QDD435 pKa = 3.79EE436 pKa = 4.62TRR438 pKa = 11.84ATYY441 pKa = 10.48GPEE444 pKa = 3.67LSNADD449 pKa = 3.41FATAVYY455 pKa = 10.26GNVLGRR461 pKa = 11.84VADD464 pKa = 3.61QAGFDD469 pKa = 3.67FWVGVLDD476 pKa = 3.54SGARR480 pKa = 11.84TRR482 pKa = 11.84DD483 pKa = 3.39QFILSVLEE491 pKa = 4.02GAKK494 pKa = 10.05AAPFEE499 pKa = 4.31GAPADD504 pKa = 4.48FVAQQIADD512 pKa = 3.99RR513 pKa = 11.84QYY515 pKa = 11.37LSTKK519 pKa = 8.83TDD521 pKa = 2.5IGAYY525 pKa = 7.99YY526 pKa = 10.69AVIRR530 pKa = 11.84GMSDD534 pKa = 2.95VTNATAAMNAFDD546 pKa = 5.09GSAAGINAAVAAIDD560 pKa = 4.91GYY562 pKa = 10.82FQAAQDD568 pKa = 3.59GDD570 pKa = 3.77TGQFLMPLVGVLDD583 pKa = 4.26DD584 pKa = 4.73PFAGMVV590 pKa = 3.18
MM1 pKa = 7.22FPEE4 pKa = 5.11DD5 pKa = 4.13SPLLAALASYY15 pKa = 10.94GSDD18 pKa = 2.69EE19 pKa = 4.37RR20 pKa = 11.84LYY22 pKa = 10.48TSTVTFRR29 pKa = 11.84FMPAGSEE36 pKa = 4.16IDD38 pKa = 4.81DD39 pKa = 4.6GDD41 pKa = 4.39PNTPNEE47 pKa = 4.14VSVGWSEE54 pKa = 5.03PNMQYY59 pKa = 10.77VRR61 pKa = 11.84GTFDD65 pKa = 3.81YY66 pKa = 10.42ISSVVDD72 pKa = 3.16LTFAEE77 pKa = 4.85TTSSDD82 pKa = 3.16ATISFYY88 pKa = 11.08EE89 pKa = 4.06VAEE92 pKa = 4.39FSDD95 pKa = 3.81DD96 pKa = 3.21TAGYY100 pKa = 8.23VTFISEE106 pKa = 4.81GISIAVIGSQFTGPVAFDD124 pKa = 3.46NDD126 pKa = 3.8YY127 pKa = 8.61TTIIHH132 pKa = 6.87EE133 pKa = 4.42IGHH136 pKa = 7.07AIGLSHH142 pKa = 6.99PHH144 pKa = 7.3DD145 pKa = 4.97GPAVLPGVTSQDD157 pKa = 3.44DD158 pKa = 3.8KK159 pKa = 12.09GDD161 pKa = 4.08LNLNTEE167 pKa = 4.33FTTRR171 pKa = 11.84MSYY174 pKa = 10.24IPGEE178 pKa = 4.41SPSNPGVDD186 pKa = 2.54IWGEE190 pKa = 4.0ANTFGAIDD198 pKa = 3.54IAALQLLYY206 pKa = 10.74GANTQTGLGDD216 pKa = 3.6TVYY219 pKa = 11.33GDD221 pKa = 3.43TRR223 pKa = 11.84DD224 pKa = 5.3LVTIWDD230 pKa = 3.53NGGFDD235 pKa = 5.18RR236 pKa = 11.84IDD238 pKa = 3.5FSSATEE244 pKa = 3.93DD245 pKa = 3.43AVIDD249 pKa = 3.85LRR251 pKa = 11.84AATLQFEE258 pKa = 4.75VGGGGYY264 pKa = 10.48LSYY267 pKa = 10.64ISSRR271 pKa = 11.84NGTVADD277 pKa = 3.61GGYY280 pKa = 10.23AIAYY284 pKa = 8.49GVEE287 pKa = 3.98IEE289 pKa = 4.23NAMGGAGADD298 pKa = 3.27MLMGNALANTFVGGGGNDD316 pKa = 3.56VIDD319 pKa = 4.65GGDD322 pKa = 3.56GRR324 pKa = 11.84DD325 pKa = 3.19TGAYY329 pKa = 9.71SGNQSSYY336 pKa = 10.58TLTLSAQTTTLADD349 pKa = 3.52RR350 pKa = 11.84RR351 pKa = 11.84GTDD354 pKa = 3.32GTDD357 pKa = 2.86TLVNMEE363 pKa = 5.26LLTFGDD369 pKa = 3.71AAVAPFDD376 pKa = 3.68LTVFAGTTGLSEE388 pKa = 4.14TQMEE392 pKa = 4.67SFIEE396 pKa = 4.25LYY398 pKa = 9.75IAYY401 pKa = 9.93FNRR404 pKa = 11.84APDD407 pKa = 3.65AVGLNFWGTAFASGTTLEE425 pKa = 4.12QMATLFTDD433 pKa = 3.29QDD435 pKa = 3.79EE436 pKa = 4.62TRR438 pKa = 11.84ATYY441 pKa = 10.48GPEE444 pKa = 3.67LSNADD449 pKa = 3.41FATAVYY455 pKa = 10.26GNVLGRR461 pKa = 11.84VADD464 pKa = 3.61QAGFDD469 pKa = 3.67FWVGVLDD476 pKa = 3.54SGARR480 pKa = 11.84TRR482 pKa = 11.84DD483 pKa = 3.39QFILSVLEE491 pKa = 4.02GAKK494 pKa = 10.05AAPFEE499 pKa = 4.31GAPADD504 pKa = 4.48FVAQQIADD512 pKa = 3.99RR513 pKa = 11.84QYY515 pKa = 11.37LSTKK519 pKa = 8.83TDD521 pKa = 2.5IGAYY525 pKa = 7.99YY526 pKa = 10.69AVIRR530 pKa = 11.84GMSDD534 pKa = 2.95VTNATAAMNAFDD546 pKa = 5.09GSAAGINAAVAAIDD560 pKa = 4.91GYY562 pKa = 10.82FQAAQDD568 pKa = 3.59GDD570 pKa = 3.77TGQFLMPLVGVLDD583 pKa = 4.26DD584 pKa = 4.73PFAGMVV590 pKa = 3.18
Molecular weight: 62.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P0CFE8|A0A6P0CFE8_9RHOB MBL fold metallo-hydrolase OS=Sulfitobacter sp. JBTF-M27 OX=2698830 GN=GV827_21190 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.34SLSAA44 pKa = 3.83
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.34SLSAA44 pKa = 3.83
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1404947 |
34 |
3221 |
313.0 |
34.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.728 ± 0.046 | 0.901 ± 0.012 |
6.258 ± 0.036 | 6.009 ± 0.035 |
3.823 ± 0.024 | 8.41 ± 0.04 |
2.063 ± 0.02 | 5.449 ± 0.027 |
3.596 ± 0.035 | 9.816 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.894 ± 0.019 | 2.835 ± 0.02 |
4.91 ± 0.026 | 3.353 ± 0.02 |
6.234 ± 0.033 | 5.324 ± 0.023 |
5.553 ± 0.027 | 7.18 ± 0.031 |
1.382 ± 0.013 | 2.281 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
