
Apis mellifera associated microvirus 44
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
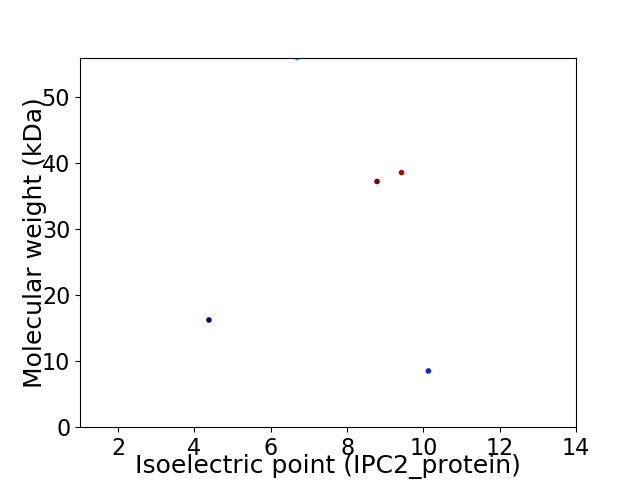
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
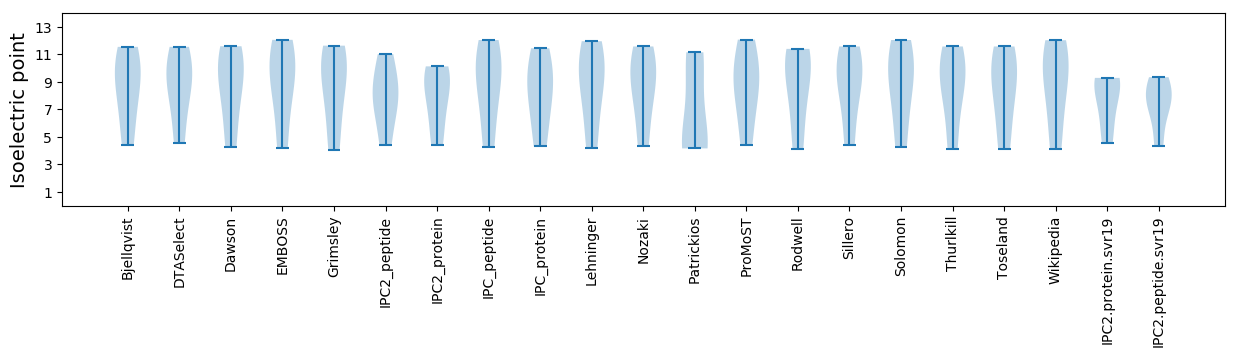
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8U598|A0A3Q8U598_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 44 OX=2494774 PE=4 SV=1
MM1 pKa = 7.44SSKK4 pKa = 10.51KK5 pKa = 9.89PSTEE9 pKa = 3.5IPFDD13 pKa = 3.36IAPRR17 pKa = 11.84APRR20 pKa = 11.84FQNLNTLGYY29 pKa = 9.95SIADD33 pKa = 3.97PTPVAPPIGYY43 pKa = 8.76QKK45 pKa = 9.64PISMVDD51 pKa = 2.88QMRR54 pKa = 11.84NLIRR58 pKa = 11.84SEE60 pKa = 3.76MLARR64 pKa = 11.84AVAAQGMEE72 pKa = 4.1TFEE75 pKa = 4.17EE76 pKa = 4.67SEE78 pKa = 4.58DD79 pKa = 3.96FDD81 pKa = 5.16VPDD84 pKa = 5.47DD85 pKa = 5.07DD86 pKa = 5.95GVLDD90 pKa = 4.1PHH92 pKa = 7.09SPWEE96 pKa = 4.03NDD98 pKa = 2.61FDD100 pKa = 4.31PPARR104 pKa = 11.84EE105 pKa = 3.54IAEE108 pKa = 4.14AVGDD112 pKa = 3.92ARR114 pKa = 11.84QRR116 pKa = 11.84PPMPAEE122 pKa = 4.38GGGGGAAPAEE132 pKa = 4.28QPKK135 pKa = 9.45PASPAPAPEE144 pKa = 4.58APPAKK149 pKa = 9.7PALPP153 pKa = 4.09
MM1 pKa = 7.44SSKK4 pKa = 10.51KK5 pKa = 9.89PSTEE9 pKa = 3.5IPFDD13 pKa = 3.36IAPRR17 pKa = 11.84APRR20 pKa = 11.84FQNLNTLGYY29 pKa = 9.95SIADD33 pKa = 3.97PTPVAPPIGYY43 pKa = 8.76QKK45 pKa = 9.64PISMVDD51 pKa = 2.88QMRR54 pKa = 11.84NLIRR58 pKa = 11.84SEE60 pKa = 3.76MLARR64 pKa = 11.84AVAAQGMEE72 pKa = 4.1TFEE75 pKa = 4.17EE76 pKa = 4.67SEE78 pKa = 4.58DD79 pKa = 3.96FDD81 pKa = 5.16VPDD84 pKa = 5.47DD85 pKa = 5.07DD86 pKa = 5.95GVLDD90 pKa = 4.1PHH92 pKa = 7.09SPWEE96 pKa = 4.03NDD98 pKa = 2.61FDD100 pKa = 4.31PPARR104 pKa = 11.84EE105 pKa = 3.54IAEE108 pKa = 4.14AVGDD112 pKa = 3.92ARR114 pKa = 11.84QRR116 pKa = 11.84PPMPAEE122 pKa = 4.38GGGGGAAPAEE132 pKa = 4.28QPKK135 pKa = 9.45PASPAPAPEE144 pKa = 4.58APPAKK149 pKa = 9.7PALPP153 pKa = 4.09
Molecular weight: 16.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTI2|A0A3S8UTI2_9VIRU Uncharacterized protein OS=Apis mellifera associated microvirus 44 OX=2494774 PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 10.6GGGLNGLSSSSSSDD16 pKa = 3.16CSSSSRR22 pKa = 11.84SRR24 pKa = 11.84SSSSRR29 pKa = 11.84SRR31 pKa = 11.84APNRR35 pKa = 11.84VSHH38 pKa = 5.79VRR40 pKa = 11.84CEE42 pKa = 4.1NPFVRR47 pKa = 11.84GNSVHH52 pKa = 6.25GCGRR56 pKa = 11.84CLPCRR61 pKa = 11.84INRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84LWKK69 pKa = 10.4VRR71 pKa = 11.84LGLEE75 pKa = 4.24ALLHH79 pKa = 5.36SQSAFVTLTYY89 pKa = 10.72ADD91 pKa = 3.89EE92 pKa = 4.42NLPVAKK98 pKa = 10.11SSKK101 pKa = 10.61GSLATLDD108 pKa = 4.0PVDD111 pKa = 3.94LRR113 pKa = 11.84NWLKK117 pKa = 10.64RR118 pKa = 11.84LRR120 pKa = 11.84KK121 pKa = 9.6AISPSSIRR129 pKa = 11.84YY130 pKa = 7.64YY131 pKa = 11.24AVGEE135 pKa = 4.2YY136 pKa = 10.63GDD138 pKa = 4.16EE139 pKa = 4.0SWRR142 pKa = 11.84PHH144 pKa = 4.14YY145 pKa = 10.57HH146 pKa = 6.46LALFGYY152 pKa = 7.51PRR154 pKa = 11.84CSTLQTTYY162 pKa = 11.01RR163 pKa = 11.84LRR165 pKa = 11.84ASTSEE170 pKa = 3.97KK171 pKa = 10.34PKK173 pKa = 10.95CCDD176 pKa = 2.82HH177 pKa = 6.88CRR179 pKa = 11.84LIHH182 pKa = 6.93DD183 pKa = 3.53SWGLGRR189 pKa = 11.84VQVAEE194 pKa = 4.75LNDD197 pKa = 3.43ASAGYY202 pKa = 9.38IAGYY206 pKa = 9.4IEE208 pKa = 4.63KK209 pKa = 10.41KK210 pKa = 9.06LTRR213 pKa = 11.84TDD215 pKa = 3.55DD216 pKa = 3.37PRR218 pKa = 11.84LDD220 pKa = 3.79GRR222 pKa = 11.84WPEE225 pKa = 4.01FARR228 pKa = 11.84MSLRR232 pKa = 11.84PGIGAGFMPHH242 pKa = 6.44YY243 pKa = 9.92FQALLEE249 pKa = 4.58HH250 pKa = 6.64NLDD253 pKa = 3.63GMADD257 pKa = 3.55VPSAIRR263 pKa = 11.84IGDD266 pKa = 3.57KK267 pKa = 10.41VVPLGRR273 pKa = 11.84YY274 pKa = 7.46LTSQLRR280 pKa = 11.84KK281 pKa = 10.31LNGKK285 pKa = 9.12DD286 pKa = 3.12GKK288 pKa = 10.78APPEE292 pKa = 3.7TLKK295 pKa = 11.09AVFEE299 pKa = 4.44EE300 pKa = 4.69KK301 pKa = 10.69VLPLLLAARR310 pKa = 11.84TSADD314 pKa = 3.31APSLKK319 pKa = 10.49EE320 pKa = 4.05QVLQKK325 pKa = 10.86NRR327 pKa = 11.84GARR330 pKa = 11.84SRR332 pKa = 11.84LLSLNKK338 pKa = 9.25IHH340 pKa = 7.12RR341 pKa = 11.84QRR343 pKa = 11.84KK344 pKa = 7.8HH345 pKa = 4.65LL346 pKa = 4.02
MM1 pKa = 7.65KK2 pKa = 10.6GGGLNGLSSSSSSDD16 pKa = 3.16CSSSSRR22 pKa = 11.84SRR24 pKa = 11.84SSSSRR29 pKa = 11.84SRR31 pKa = 11.84APNRR35 pKa = 11.84VSHH38 pKa = 5.79VRR40 pKa = 11.84CEE42 pKa = 4.1NPFVRR47 pKa = 11.84GNSVHH52 pKa = 6.25GCGRR56 pKa = 11.84CLPCRR61 pKa = 11.84INRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84LWKK69 pKa = 10.4VRR71 pKa = 11.84LGLEE75 pKa = 4.24ALLHH79 pKa = 5.36SQSAFVTLTYY89 pKa = 10.72ADD91 pKa = 3.89EE92 pKa = 4.42NLPVAKK98 pKa = 10.11SSKK101 pKa = 10.61GSLATLDD108 pKa = 4.0PVDD111 pKa = 3.94LRR113 pKa = 11.84NWLKK117 pKa = 10.64RR118 pKa = 11.84LRR120 pKa = 11.84KK121 pKa = 9.6AISPSSIRR129 pKa = 11.84YY130 pKa = 7.64YY131 pKa = 11.24AVGEE135 pKa = 4.2YY136 pKa = 10.63GDD138 pKa = 4.16EE139 pKa = 4.0SWRR142 pKa = 11.84PHH144 pKa = 4.14YY145 pKa = 10.57HH146 pKa = 6.46LALFGYY152 pKa = 7.51PRR154 pKa = 11.84CSTLQTTYY162 pKa = 11.01RR163 pKa = 11.84LRR165 pKa = 11.84ASTSEE170 pKa = 3.97KK171 pKa = 10.34PKK173 pKa = 10.95CCDD176 pKa = 2.82HH177 pKa = 6.88CRR179 pKa = 11.84LIHH182 pKa = 6.93DD183 pKa = 3.53SWGLGRR189 pKa = 11.84VQVAEE194 pKa = 4.75LNDD197 pKa = 3.43ASAGYY202 pKa = 9.38IAGYY206 pKa = 9.4IEE208 pKa = 4.63KK209 pKa = 10.41KK210 pKa = 9.06LTRR213 pKa = 11.84TDD215 pKa = 3.55DD216 pKa = 3.37PRR218 pKa = 11.84LDD220 pKa = 3.79GRR222 pKa = 11.84WPEE225 pKa = 4.01FARR228 pKa = 11.84MSLRR232 pKa = 11.84PGIGAGFMPHH242 pKa = 6.44YY243 pKa = 9.92FQALLEE249 pKa = 4.58HH250 pKa = 6.64NLDD253 pKa = 3.63GMADD257 pKa = 3.55VPSAIRR263 pKa = 11.84IGDD266 pKa = 3.57KK267 pKa = 10.41VVPLGRR273 pKa = 11.84YY274 pKa = 7.46LTSQLRR280 pKa = 11.84KK281 pKa = 10.31LNGKK285 pKa = 9.12DD286 pKa = 3.12GKK288 pKa = 10.78APPEE292 pKa = 3.7TLKK295 pKa = 11.09AVFEE299 pKa = 4.44EE300 pKa = 4.69KK301 pKa = 10.69VLPLLLAARR310 pKa = 11.84TSADD314 pKa = 3.31APSLKK319 pKa = 10.49EE320 pKa = 4.05QVLQKK325 pKa = 10.86NRR327 pKa = 11.84GARR330 pKa = 11.84SRR332 pKa = 11.84LLSLNKK338 pKa = 9.25IHH340 pKa = 7.12RR341 pKa = 11.84QRR343 pKa = 11.84KK344 pKa = 7.8HH345 pKa = 4.65LL346 pKa = 4.02
Molecular weight: 38.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1420 |
75 |
499 |
284.0 |
31.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.0 ± 1.423 | 0.845 ± 0.551 |
5.07 ± 0.575 | 4.507 ± 0.509 |
3.732 ± 0.68 | 8.099 ± 0.717 |
2.394 ± 0.334 | 4.085 ± 0.742 |
4.085 ± 0.737 | 7.746 ± 1.479 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.746 ± 0.613 | 4.366 ± 0.569 |
7.324 ± 1.679 | 4.296 ± 0.872 |
7.746 ± 1.367 | 8.028 ± 1.071 |
4.93 ± 1.017 | 5.211 ± 0.462 |
1.338 ± 0.278 | 3.451 ± 0.513 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
