
Maize mosaic virus (isolate Maize/United States/Reed/2005) (MMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus; Maize mosaic nucleorhabdovirus
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
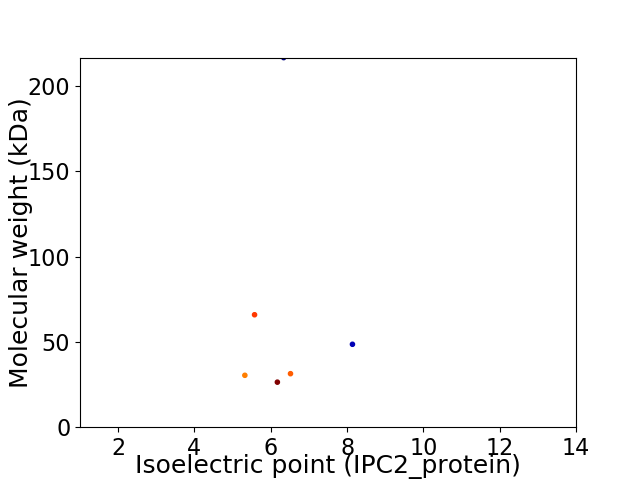
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q6E0X1|NCAP_MMVR Nucleoprotein OS=Maize mosaic virus (isolate Maize/United States/Reed/2005) OX=928305 GN=N PE=3 SV=1
MM1 pKa = 7.09NRR3 pKa = 11.84YY4 pKa = 8.65SRR6 pKa = 11.84RR7 pKa = 11.84SRR9 pKa = 11.84HH10 pKa = 5.9PNPPVPNQEE19 pKa = 4.02EE20 pKa = 4.56PEE22 pKa = 4.1RR23 pKa = 11.84DD24 pKa = 3.77PNHH27 pKa = 6.96IDD29 pKa = 3.82QDD31 pKa = 4.33LADD34 pKa = 4.55LAQPLVLKK42 pKa = 10.04EE43 pKa = 3.94RR44 pKa = 11.84HH45 pKa = 5.76AVMAPTQPSLSDD57 pKa = 3.5VINEE61 pKa = 3.77EE62 pKa = 3.77RR63 pKa = 11.84QAPITFGNPPEE74 pKa = 4.23VMANARR80 pKa = 11.84LSALGYY86 pKa = 10.87DD87 pKa = 3.43NLTEE91 pKa = 3.99RR92 pKa = 11.84EE93 pKa = 3.96KK94 pKa = 10.94RR95 pKa = 11.84ILAVGVRR102 pKa = 11.84CGEE105 pKa = 3.87AAKK108 pKa = 10.19DD109 pKa = 3.77YY110 pKa = 11.32HH111 pKa = 7.73SLTTTKK117 pKa = 10.38KK118 pKa = 9.62WIEE121 pKa = 4.51DD122 pKa = 3.56EE123 pKa = 5.29LKK125 pKa = 10.6SQMVALASSTRR136 pKa = 11.84TLTEE140 pKa = 3.65AASLHH145 pKa = 4.98TTFAMLHH152 pKa = 5.7SPSIKK157 pKa = 9.96RR158 pKa = 11.84KK159 pKa = 9.49AEE161 pKa = 3.97AMSHH165 pKa = 5.99ISQGEE170 pKa = 3.82EE171 pKa = 3.77SIDD174 pKa = 3.2ISKK177 pKa = 10.72LNKK180 pKa = 9.61TGMEE184 pKa = 4.85DD185 pKa = 2.28IWVAMEE191 pKa = 4.33SEE193 pKa = 4.4SKK195 pKa = 10.65EE196 pKa = 4.24DD197 pKa = 4.61AVDD200 pKa = 3.52TYY202 pKa = 11.31LRR204 pKa = 11.84NILEE208 pKa = 4.03VDD210 pKa = 3.32PTQFYY215 pKa = 10.83AIDD218 pKa = 3.38GWGLYY223 pKa = 10.52LDD225 pKa = 6.52FIPTWHH231 pKa = 6.76YY232 pKa = 10.4IAAGKK237 pKa = 10.29NSAQFKK243 pKa = 9.53TSYY246 pKa = 10.66ADD248 pKa = 3.66EE249 pKa = 4.34IVEE252 pKa = 3.77QRR254 pKa = 11.84AAFEE258 pKa = 4.31RR259 pKa = 11.84VLSKK263 pKa = 10.64RR264 pKa = 11.84PRR266 pKa = 11.84VEE268 pKa = 3.33II269 pKa = 4.27
MM1 pKa = 7.09NRR3 pKa = 11.84YY4 pKa = 8.65SRR6 pKa = 11.84RR7 pKa = 11.84SRR9 pKa = 11.84HH10 pKa = 5.9PNPPVPNQEE19 pKa = 4.02EE20 pKa = 4.56PEE22 pKa = 4.1RR23 pKa = 11.84DD24 pKa = 3.77PNHH27 pKa = 6.96IDD29 pKa = 3.82QDD31 pKa = 4.33LADD34 pKa = 4.55LAQPLVLKK42 pKa = 10.04EE43 pKa = 3.94RR44 pKa = 11.84HH45 pKa = 5.76AVMAPTQPSLSDD57 pKa = 3.5VINEE61 pKa = 3.77EE62 pKa = 3.77RR63 pKa = 11.84QAPITFGNPPEE74 pKa = 4.23VMANARR80 pKa = 11.84LSALGYY86 pKa = 10.87DD87 pKa = 3.43NLTEE91 pKa = 3.99RR92 pKa = 11.84EE93 pKa = 3.96KK94 pKa = 10.94RR95 pKa = 11.84ILAVGVRR102 pKa = 11.84CGEE105 pKa = 3.87AAKK108 pKa = 10.19DD109 pKa = 3.77YY110 pKa = 11.32HH111 pKa = 7.73SLTTTKK117 pKa = 10.38KK118 pKa = 9.62WIEE121 pKa = 4.51DD122 pKa = 3.56EE123 pKa = 5.29LKK125 pKa = 10.6SQMVALASSTRR136 pKa = 11.84TLTEE140 pKa = 3.65AASLHH145 pKa = 4.98TTFAMLHH152 pKa = 5.7SPSIKK157 pKa = 9.96RR158 pKa = 11.84KK159 pKa = 9.49AEE161 pKa = 3.97AMSHH165 pKa = 5.99ISQGEE170 pKa = 3.82EE171 pKa = 3.77SIDD174 pKa = 3.2ISKK177 pKa = 10.72LNKK180 pKa = 9.61TGMEE184 pKa = 4.85DD185 pKa = 2.28IWVAMEE191 pKa = 4.33SEE193 pKa = 4.4SKK195 pKa = 10.65EE196 pKa = 4.24DD197 pKa = 4.61AVDD200 pKa = 3.52TYY202 pKa = 11.31LRR204 pKa = 11.84NILEE208 pKa = 4.03VDD210 pKa = 3.32PTQFYY215 pKa = 10.83AIDD218 pKa = 3.38GWGLYY223 pKa = 10.52LDD225 pKa = 6.52FIPTWHH231 pKa = 6.76YY232 pKa = 10.4IAAGKK237 pKa = 10.29NSAQFKK243 pKa = 9.53TSYY246 pKa = 10.66ADD248 pKa = 3.66EE249 pKa = 4.34IVEE252 pKa = 3.77QRR254 pKa = 11.84AAFEE258 pKa = 4.31RR259 pKa = 11.84VLSKK263 pKa = 10.64RR264 pKa = 11.84PRR266 pKa = 11.84VEE268 pKa = 3.33II269 pKa = 4.27
Molecular weight: 30.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q6E0X1|NCAP_MMVR Nucleoprotein OS=Maize mosaic virus (isolate Maize/United States/Reed/2005) OX=928305 GN=N PE=3 SV=1
MM1 pKa = 7.59ANINIPDD8 pKa = 4.29DD9 pKa = 4.02LVSKK13 pKa = 10.69YY14 pKa = 10.91SEE16 pKa = 4.23DD17 pKa = 4.61VKK19 pKa = 11.44DD20 pKa = 3.82LTQKK24 pKa = 10.77AGSIPSSKK32 pKa = 10.92SLIPQTAFTPDD43 pKa = 3.01EE44 pKa = 4.28LRR46 pKa = 11.84RR47 pKa = 11.84SLKK50 pKa = 9.76FWQVTTKK57 pKa = 10.52VAATVASDD65 pKa = 3.2WTNLATALTNGTFSATHH82 pKa = 6.31LRR84 pKa = 11.84TLCEE88 pKa = 3.55LAFNLKK94 pKa = 10.47APTGTGTIFIHH105 pKa = 7.06EE106 pKa = 4.68IADD109 pKa = 3.54PWKK112 pKa = 10.86GCIDD116 pKa = 3.72ANAPDD121 pKa = 4.31TVAIPATDD129 pKa = 3.26SDD131 pKa = 4.6VNMSTVSSGSTASGTEE147 pKa = 3.83DD148 pKa = 4.7AEE150 pKa = 4.62VKK152 pKa = 10.0MKK154 pKa = 10.71AISFLCCTLLRR165 pKa = 11.84LSVKK169 pKa = 9.82EE170 pKa = 4.39PSHH173 pKa = 7.67IMQAINSIRR182 pKa = 11.84QRR184 pKa = 11.84FGSLYY189 pKa = 10.06GVSSPTLNSVTFNRR203 pKa = 11.84SQLSRR208 pKa = 11.84IKK210 pKa = 10.5QGIEE214 pKa = 3.9TYY216 pKa = 9.8PSARR220 pKa = 11.84GTVFYY225 pKa = 8.14YY226 pKa = 10.24TRR228 pKa = 11.84YY229 pKa = 10.36ADD231 pKa = 3.44ATHH234 pKa = 7.11GYY236 pKa = 5.28TTKK239 pKa = 10.58EE240 pKa = 3.56YY241 pKa = 10.42GICRR245 pKa = 11.84FLLFQHH251 pKa = 6.78LEE253 pKa = 4.08LEE255 pKa = 4.52GMHH258 pKa = 6.77IYY260 pKa = 11.04KK261 pKa = 9.5MLLALLGEE269 pKa = 4.66WSTVPIGLLLTWIRR283 pKa = 11.84NPRR286 pKa = 11.84SKK288 pKa = 10.72LAINEE293 pKa = 3.63IVKK296 pKa = 9.9IVKK299 pKa = 9.79EE300 pKa = 3.85LDD302 pKa = 3.21KK303 pKa = 11.59PEE305 pKa = 3.89VDD307 pKa = 4.73KK308 pKa = 11.1GWKK311 pKa = 7.57YY312 pKa = 11.42ARR314 pKa = 11.84MVNNTFFLDD323 pKa = 3.98LSSRR327 pKa = 11.84RR328 pKa = 11.84NTYY331 pKa = 10.14LCAVLASLNKK341 pKa = 10.06KK342 pKa = 9.32NVPQGSGEE350 pKa = 4.15YY351 pKa = 10.49ADD353 pKa = 3.88PTNIAVIKK361 pKa = 11.17NMDD364 pKa = 3.56QSVKK368 pKa = 10.55SQVATDD374 pKa = 3.26VTLIEE379 pKa = 4.83RR380 pKa = 11.84IYY382 pKa = 10.98EE383 pKa = 4.16KK384 pKa = 11.1YY385 pKa = 10.35LVSAGGDD392 pKa = 3.56EE393 pKa = 5.15AGTAYY398 pKa = 10.55ALSRR402 pKa = 11.84GVKK405 pKa = 9.66RR406 pKa = 11.84SSPPSQEE413 pKa = 4.14GGQSGQSGGTPMEE426 pKa = 3.86VDD428 pKa = 3.55GASGRR433 pKa = 11.84GAAGPAPKK441 pKa = 8.91KK442 pKa = 8.96TRR444 pKa = 11.84SGLL447 pKa = 3.57
MM1 pKa = 7.59ANINIPDD8 pKa = 4.29DD9 pKa = 4.02LVSKK13 pKa = 10.69YY14 pKa = 10.91SEE16 pKa = 4.23DD17 pKa = 4.61VKK19 pKa = 11.44DD20 pKa = 3.82LTQKK24 pKa = 10.77AGSIPSSKK32 pKa = 10.92SLIPQTAFTPDD43 pKa = 3.01EE44 pKa = 4.28LRR46 pKa = 11.84RR47 pKa = 11.84SLKK50 pKa = 9.76FWQVTTKK57 pKa = 10.52VAATVASDD65 pKa = 3.2WTNLATALTNGTFSATHH82 pKa = 6.31LRR84 pKa = 11.84TLCEE88 pKa = 3.55LAFNLKK94 pKa = 10.47APTGTGTIFIHH105 pKa = 7.06EE106 pKa = 4.68IADD109 pKa = 3.54PWKK112 pKa = 10.86GCIDD116 pKa = 3.72ANAPDD121 pKa = 4.31TVAIPATDD129 pKa = 3.26SDD131 pKa = 4.6VNMSTVSSGSTASGTEE147 pKa = 3.83DD148 pKa = 4.7AEE150 pKa = 4.62VKK152 pKa = 10.0MKK154 pKa = 10.71AISFLCCTLLRR165 pKa = 11.84LSVKK169 pKa = 9.82EE170 pKa = 4.39PSHH173 pKa = 7.67IMQAINSIRR182 pKa = 11.84QRR184 pKa = 11.84FGSLYY189 pKa = 10.06GVSSPTLNSVTFNRR203 pKa = 11.84SQLSRR208 pKa = 11.84IKK210 pKa = 10.5QGIEE214 pKa = 3.9TYY216 pKa = 9.8PSARR220 pKa = 11.84GTVFYY225 pKa = 8.14YY226 pKa = 10.24TRR228 pKa = 11.84YY229 pKa = 10.36ADD231 pKa = 3.44ATHH234 pKa = 7.11GYY236 pKa = 5.28TTKK239 pKa = 10.58EE240 pKa = 3.56YY241 pKa = 10.42GICRR245 pKa = 11.84FLLFQHH251 pKa = 6.78LEE253 pKa = 4.08LEE255 pKa = 4.52GMHH258 pKa = 6.77IYY260 pKa = 11.04KK261 pKa = 9.5MLLALLGEE269 pKa = 4.66WSTVPIGLLLTWIRR283 pKa = 11.84NPRR286 pKa = 11.84SKK288 pKa = 10.72LAINEE293 pKa = 3.63IVKK296 pKa = 9.9IVKK299 pKa = 9.79EE300 pKa = 3.85LDD302 pKa = 3.21KK303 pKa = 11.59PEE305 pKa = 3.89VDD307 pKa = 4.73KK308 pKa = 11.1GWKK311 pKa = 7.57YY312 pKa = 11.42ARR314 pKa = 11.84MVNNTFFLDD323 pKa = 3.98LSSRR327 pKa = 11.84RR328 pKa = 11.84NTYY331 pKa = 10.14LCAVLASLNKK341 pKa = 10.06KK342 pKa = 9.32NVPQGSGEE350 pKa = 4.15YY351 pKa = 10.49ADD353 pKa = 3.88PTNIAVIKK361 pKa = 11.17NMDD364 pKa = 3.56QSVKK368 pKa = 10.55SQVATDD374 pKa = 3.26VTLIEE379 pKa = 4.83RR380 pKa = 11.84IYY382 pKa = 10.98EE383 pKa = 4.16KK384 pKa = 11.1YY385 pKa = 10.35LVSAGGDD392 pKa = 3.56EE393 pKa = 5.15AGTAYY398 pKa = 10.55ALSRR402 pKa = 11.84GVKK405 pKa = 9.66RR406 pKa = 11.84SSPPSQEE413 pKa = 4.14GGQSGQSGGTPMEE426 pKa = 3.86VDD428 pKa = 3.55GASGRR433 pKa = 11.84GAAGPAPKK441 pKa = 8.91KK442 pKa = 8.96TRR444 pKa = 11.84SGLL447 pKa = 3.57
Molecular weight: 48.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3750 |
235 |
1922 |
625.0 |
69.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.187 ± 0.685 | 1.627 ± 0.236 |
5.6 ± 0.344 | 5.68 ± 0.478 |
3.04 ± 0.204 | 6.427 ± 0.458 |
2.667 ± 0.223 | 7.093 ± 0.45 |
5.253 ± 0.186 | 10.16 ± 0.741 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.8 ± 0.242 | 3.867 ± 0.265 |
4.453 ± 0.426 | 2.747 ± 0.234 |
5.44 ± 0.598 | 8.773 ± 0.798 |
7.013 ± 0.558 | 5.893 ± 0.239 |
1.787 ± 0.331 | 3.413 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
