
Lake Sarah-associated circular virus-40
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.09
Get precalculated fractions of proteins
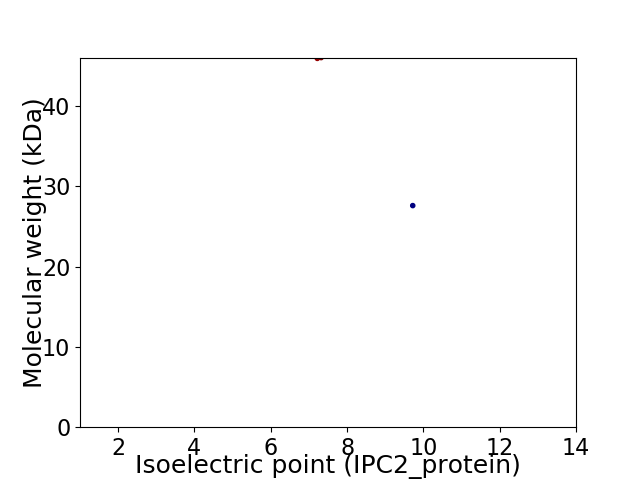
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
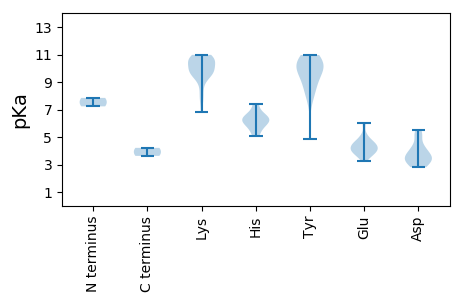
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQQ2|A0A140AQQ2_9VIRU Coat protein OS=Lake Sarah-associated circular virus-40 OX=1685769 PE=4 SV=1
MM1 pKa = 7.85EE2 pKa = 4.36EE3 pKa = 3.92TTTEE7 pKa = 3.86KK8 pKa = 10.88SVISTGGGPQSSRR21 pKa = 11.84TWCGTLNNYY30 pKa = 8.68SQGEE34 pKa = 4.32YY35 pKa = 10.55EE36 pKa = 4.66FLVQEE41 pKa = 4.25FNKK44 pKa = 9.72VAEE47 pKa = 3.86YY48 pKa = 9.79WIIGKK53 pKa = 9.63EE54 pKa = 3.9KK55 pKa = 9.37GQEE58 pKa = 4.22GTPHH62 pKa = 6.59LQMACSFTRR71 pKa = 11.84AQKK74 pKa = 10.82LNYY77 pKa = 9.47LKK79 pKa = 10.85QNLSVRR85 pKa = 11.84AHH87 pKa = 5.65WEE89 pKa = 3.79ITKK92 pKa = 8.89GTLDD96 pKa = 3.48QNRR99 pKa = 11.84VYY101 pKa = 10.85CMKK104 pKa = 10.63EE105 pKa = 3.28GDD107 pKa = 4.03YY108 pKa = 10.97TEE110 pKa = 5.95GGAPPVKK117 pKa = 10.24KK118 pKa = 9.99QGKK121 pKa = 7.51RR122 pKa = 11.84TDD124 pKa = 2.82IEE126 pKa = 4.34MAVEE130 pKa = 4.02TLKK133 pKa = 10.63ATGGDD138 pKa = 3.79VQEE141 pKa = 4.65MALTHH146 pKa = 6.28PEE148 pKa = 4.0AYY150 pKa = 9.77VKK152 pKa = 10.86YY153 pKa = 10.22SSNFEE158 pKa = 4.13KK159 pKa = 10.71LAQRR163 pKa = 11.84YY164 pKa = 4.84QKK166 pKa = 10.87KK167 pKa = 8.66MRR169 pKa = 11.84SSGSGYY175 pKa = 8.59VHH177 pKa = 7.4DD178 pKa = 5.52RR179 pKa = 11.84RR180 pKa = 11.84VIWIYY185 pKa = 10.95GPSGTGKK192 pKa = 7.95TRR194 pKa = 11.84YY195 pKa = 8.23VHH197 pKa = 6.25EE198 pKa = 5.27KK199 pKa = 10.65EE200 pKa = 5.01LISNLWISGRR210 pKa = 11.84DD211 pKa = 3.6LRR213 pKa = 11.84WWQSYY218 pKa = 10.77DD219 pKa = 3.36MEE221 pKa = 4.74PAVLFDD227 pKa = 4.23DD228 pKa = 5.23FRR230 pKa = 11.84GDD232 pKa = 3.03FCKK235 pKa = 10.58YY236 pKa = 10.31HH237 pKa = 6.18EE238 pKa = 5.03LLRR241 pKa = 11.84ILDD244 pKa = 4.48RR245 pKa = 11.84YY246 pKa = 9.59QCTVEE251 pKa = 4.07NKK253 pKa = 9.86GGSVSLQNVKK263 pKa = 10.47RR264 pKa = 11.84IWITSCFHH272 pKa = 6.47PRR274 pKa = 11.84HH275 pKa = 5.97VYY277 pKa = 8.23DD278 pKa = 3.2TRR280 pKa = 11.84EE281 pKa = 4.65DD282 pKa = 3.42IEE284 pKa = 4.08QLIRR288 pKa = 11.84RR289 pKa = 11.84IDD291 pKa = 4.1KK292 pKa = 9.5IWHH295 pKa = 5.1FTSTTKK301 pKa = 10.61TDD303 pKa = 3.24VTNEE307 pKa = 3.53KK308 pKa = 9.81EE309 pKa = 4.44YY310 pKa = 10.34PGKK313 pKa = 9.87PGGAAPDD320 pKa = 5.2FITPLALVRR329 pKa = 11.84TKK331 pKa = 10.98AVIPEE336 pKa = 4.25SPPTPTNSRR345 pKa = 11.84PNSKK349 pKa = 10.05RR350 pKa = 11.84SSTIGTEE357 pKa = 3.51LDD359 pKa = 3.45MVISRR364 pKa = 11.84GVATQPLVVLDD375 pKa = 3.82SDD377 pKa = 4.33EE378 pKa = 4.85EE379 pKa = 4.43EE380 pKa = 4.47EE381 pKa = 5.99LPGQPDD387 pKa = 3.79SSWAASQWKK396 pKa = 9.57RR397 pKa = 11.84AVKK400 pKa = 10.29KK401 pKa = 10.94SEE403 pKa = 3.63
MM1 pKa = 7.85EE2 pKa = 4.36EE3 pKa = 3.92TTTEE7 pKa = 3.86KK8 pKa = 10.88SVISTGGGPQSSRR21 pKa = 11.84TWCGTLNNYY30 pKa = 8.68SQGEE34 pKa = 4.32YY35 pKa = 10.55EE36 pKa = 4.66FLVQEE41 pKa = 4.25FNKK44 pKa = 9.72VAEE47 pKa = 3.86YY48 pKa = 9.79WIIGKK53 pKa = 9.63EE54 pKa = 3.9KK55 pKa = 9.37GQEE58 pKa = 4.22GTPHH62 pKa = 6.59LQMACSFTRR71 pKa = 11.84AQKK74 pKa = 10.82LNYY77 pKa = 9.47LKK79 pKa = 10.85QNLSVRR85 pKa = 11.84AHH87 pKa = 5.65WEE89 pKa = 3.79ITKK92 pKa = 8.89GTLDD96 pKa = 3.48QNRR99 pKa = 11.84VYY101 pKa = 10.85CMKK104 pKa = 10.63EE105 pKa = 3.28GDD107 pKa = 4.03YY108 pKa = 10.97TEE110 pKa = 5.95GGAPPVKK117 pKa = 10.24KK118 pKa = 9.99QGKK121 pKa = 7.51RR122 pKa = 11.84TDD124 pKa = 2.82IEE126 pKa = 4.34MAVEE130 pKa = 4.02TLKK133 pKa = 10.63ATGGDD138 pKa = 3.79VQEE141 pKa = 4.65MALTHH146 pKa = 6.28PEE148 pKa = 4.0AYY150 pKa = 9.77VKK152 pKa = 10.86YY153 pKa = 10.22SSNFEE158 pKa = 4.13KK159 pKa = 10.71LAQRR163 pKa = 11.84YY164 pKa = 4.84QKK166 pKa = 10.87KK167 pKa = 8.66MRR169 pKa = 11.84SSGSGYY175 pKa = 8.59VHH177 pKa = 7.4DD178 pKa = 5.52RR179 pKa = 11.84RR180 pKa = 11.84VIWIYY185 pKa = 10.95GPSGTGKK192 pKa = 7.95TRR194 pKa = 11.84YY195 pKa = 8.23VHH197 pKa = 6.25EE198 pKa = 5.27KK199 pKa = 10.65EE200 pKa = 5.01LISNLWISGRR210 pKa = 11.84DD211 pKa = 3.6LRR213 pKa = 11.84WWQSYY218 pKa = 10.77DD219 pKa = 3.36MEE221 pKa = 4.74PAVLFDD227 pKa = 4.23DD228 pKa = 5.23FRR230 pKa = 11.84GDD232 pKa = 3.03FCKK235 pKa = 10.58YY236 pKa = 10.31HH237 pKa = 6.18EE238 pKa = 5.03LLRR241 pKa = 11.84ILDD244 pKa = 4.48RR245 pKa = 11.84YY246 pKa = 9.59QCTVEE251 pKa = 4.07NKK253 pKa = 9.86GGSVSLQNVKK263 pKa = 10.47RR264 pKa = 11.84IWITSCFHH272 pKa = 6.47PRR274 pKa = 11.84HH275 pKa = 5.97VYY277 pKa = 8.23DD278 pKa = 3.2TRR280 pKa = 11.84EE281 pKa = 4.65DD282 pKa = 3.42IEE284 pKa = 4.08QLIRR288 pKa = 11.84RR289 pKa = 11.84IDD291 pKa = 4.1KK292 pKa = 9.5IWHH295 pKa = 5.1FTSTTKK301 pKa = 10.61TDD303 pKa = 3.24VTNEE307 pKa = 3.53KK308 pKa = 9.81EE309 pKa = 4.44YY310 pKa = 10.34PGKK313 pKa = 9.87PGGAAPDD320 pKa = 5.2FITPLALVRR329 pKa = 11.84TKK331 pKa = 10.98AVIPEE336 pKa = 4.25SPPTPTNSRR345 pKa = 11.84PNSKK349 pKa = 10.05RR350 pKa = 11.84SSTIGTEE357 pKa = 3.51LDD359 pKa = 3.45MVISRR364 pKa = 11.84GVATQPLVVLDD375 pKa = 3.82SDD377 pKa = 4.33EE378 pKa = 4.85EE379 pKa = 4.43EE380 pKa = 4.47EE381 pKa = 5.99LPGQPDD387 pKa = 3.79SSWAASQWKK396 pKa = 9.57RR397 pKa = 11.84AVKK400 pKa = 10.29KK401 pKa = 10.94SEE403 pKa = 3.63
Molecular weight: 45.95 kDa
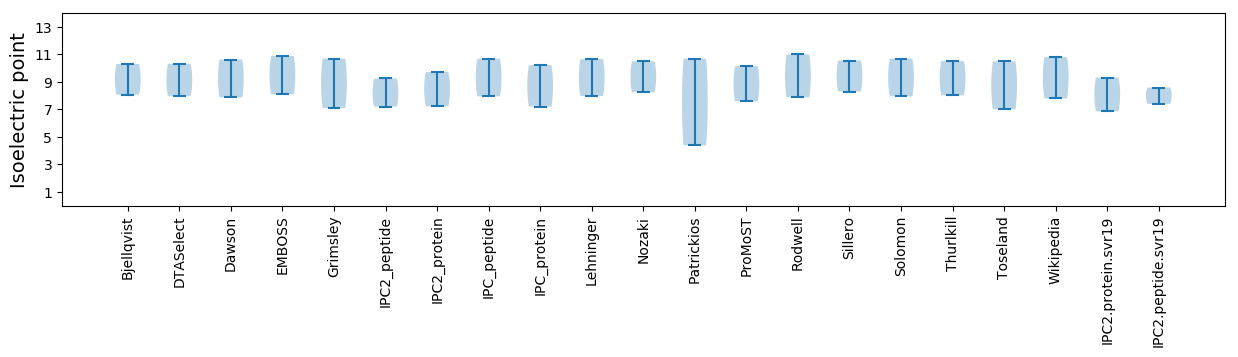
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQQ2|A0A140AQQ2_9VIRU Coat protein OS=Lake Sarah-associated circular virus-40 OX=1685769 PE=4 SV=1
MM1 pKa = 7.23PGKK4 pKa = 9.84RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.82YY8 pKa = 10.17SLGSKK13 pKa = 9.32AQIRR17 pKa = 11.84KK18 pKa = 6.8MARR21 pKa = 11.84AYY23 pKa = 9.99VRR25 pKa = 11.84KK26 pKa = 9.69KK27 pKa = 10.04YY28 pKa = 9.37RR29 pKa = 11.84TGGVPVTRR37 pKa = 11.84GAYY40 pKa = 8.96NMRR43 pKa = 11.84MPSEE47 pKa = 4.57LKK49 pKa = 10.59LSQNQTLGLTTLQNTPTIANNQLILLNSIAQGTDD83 pKa = 2.8YY84 pKa = 7.56TTRR87 pKa = 11.84IGRR90 pKa = 11.84TCLATSLQWNLEE102 pKa = 4.05LFQAPGTTTSNNQIWVRR119 pKa = 11.84AMVFCDD125 pKa = 3.37RR126 pKa = 11.84QPNGVEE132 pKa = 3.55PSVFSDD138 pKa = 3.51VLDD141 pKa = 3.49STFRR145 pKa = 11.84TEE147 pKa = 3.62QAFRR151 pKa = 11.84NLSNRR156 pKa = 11.84DD157 pKa = 2.94RR158 pKa = 11.84FVIVANKK165 pKa = 8.07VWSWSDD171 pKa = 2.81WNTLQPEE178 pKa = 4.49SLGCAKK184 pKa = 10.74NGGPKK189 pKa = 9.49RR190 pKa = 11.84WAKK193 pKa = 9.75YY194 pKa = 10.02KK195 pKa = 10.38KK196 pKa = 10.4LNVEE200 pKa = 3.84KK201 pKa = 9.43TVYY204 pKa = 10.29SGTSGGITNVQSCALFFAIWLVNTSGAVQANFAYY238 pKa = 10.43RR239 pKa = 11.84LRR241 pKa = 11.84FVDD244 pKa = 5.09FF245 pKa = 4.21
MM1 pKa = 7.23PGKK4 pKa = 9.84RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.82YY8 pKa = 10.17SLGSKK13 pKa = 9.32AQIRR17 pKa = 11.84KK18 pKa = 6.8MARR21 pKa = 11.84AYY23 pKa = 9.99VRR25 pKa = 11.84KK26 pKa = 9.69KK27 pKa = 10.04YY28 pKa = 9.37RR29 pKa = 11.84TGGVPVTRR37 pKa = 11.84GAYY40 pKa = 8.96NMRR43 pKa = 11.84MPSEE47 pKa = 4.57LKK49 pKa = 10.59LSQNQTLGLTTLQNTPTIANNQLILLNSIAQGTDD83 pKa = 2.8YY84 pKa = 7.56TTRR87 pKa = 11.84IGRR90 pKa = 11.84TCLATSLQWNLEE102 pKa = 4.05LFQAPGTTTSNNQIWVRR119 pKa = 11.84AMVFCDD125 pKa = 3.37RR126 pKa = 11.84QPNGVEE132 pKa = 3.55PSVFSDD138 pKa = 3.51VLDD141 pKa = 3.49STFRR145 pKa = 11.84TEE147 pKa = 3.62QAFRR151 pKa = 11.84NLSNRR156 pKa = 11.84DD157 pKa = 2.94RR158 pKa = 11.84FVIVANKK165 pKa = 8.07VWSWSDD171 pKa = 2.81WNTLQPEE178 pKa = 4.49SLGCAKK184 pKa = 10.74NGGPKK189 pKa = 9.49RR190 pKa = 11.84WAKK193 pKa = 9.75YY194 pKa = 10.02KK195 pKa = 10.38KK196 pKa = 10.4LNVEE200 pKa = 3.84KK201 pKa = 9.43TVYY204 pKa = 10.29SGTSGGITNVQSCALFFAIWLVNTSGAVQANFAYY238 pKa = 10.43RR239 pKa = 11.84LRR241 pKa = 11.84FVDD244 pKa = 5.09FF245 pKa = 4.21
Molecular weight: 27.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
648 |
245 |
403 |
324.0 |
36.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.71 ± 0.934 | 1.543 ± 0.051 |
4.167 ± 0.747 | 6.019 ± 2.036 |
3.241 ± 0.712 | 7.253 ± 0.179 |
1.389 ± 0.792 | 4.475 ± 0.457 |
6.79 ± 0.614 | 7.099 ± 0.84 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.006 ± 0.02 | 4.938 ± 1.607 |
4.475 ± 0.457 | 5.093 ± 0.355 |
6.79 ± 0.55 | 7.562 ± 0.123 |
8.333 ± 0.369 | 6.481 ± 0.261 |
2.778 ± 0.045 | 3.858 ± 0.338 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
