
Adelphocoris suturalis virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
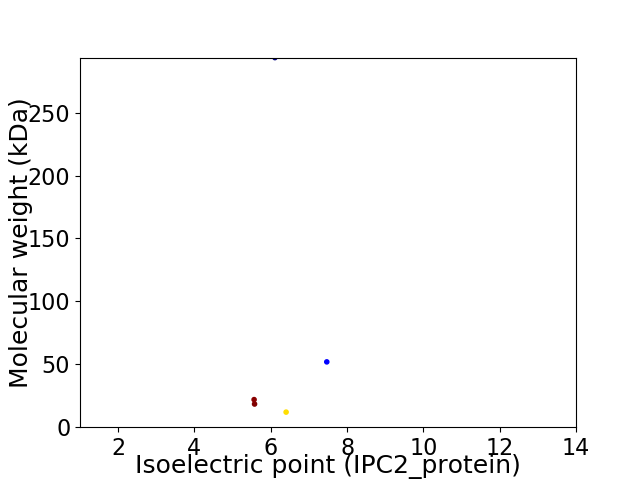
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L6YQ04|A0A1L6YQ04_9VIRU ORF4 OS=Adelphocoris suturalis virus OX=1930920 PE=4 SV=1
MM1 pKa = 7.54AAALLEE7 pKa = 4.29AAEE10 pKa = 4.15IAAEE14 pKa = 4.18IIEE17 pKa = 4.22AAKK20 pKa = 10.33LAEE23 pKa = 4.03EE24 pKa = 5.05LGTGAYY30 pKa = 10.26DD31 pKa = 4.17LATSAYY37 pKa = 9.27RR38 pKa = 11.84GLKK41 pKa = 10.15RR42 pKa = 11.84IFGTVTQRR50 pKa = 11.84YY51 pKa = 8.38EE52 pKa = 3.67PVTNFRR58 pKa = 11.84FARR61 pKa = 11.84SFTFIEE67 pKa = 4.34LPRR70 pKa = 11.84LQRR73 pKa = 11.84VLLRR77 pKa = 11.84MQNINYY83 pKa = 9.02SLYY86 pKa = 10.49YY87 pKa = 8.16EE88 pKa = 4.17RR89 pKa = 11.84EE90 pKa = 4.04YY91 pKa = 11.51ARR93 pKa = 11.84SLVSSITHH101 pKa = 5.42DD102 pKa = 2.77HH103 pKa = 6.17SMNFFVDD110 pKa = 5.08GIFRR114 pKa = 11.84EE115 pKa = 4.3DD116 pKa = 3.48VVVVIISRR124 pKa = 11.84QPFLIPFSLLSRR136 pKa = 11.84SLSLSQHH143 pKa = 5.3PTNDD147 pKa = 2.83EE148 pKa = 4.11LTTHH152 pKa = 6.94RR153 pKa = 11.84EE154 pKa = 3.71LYY156 pKa = 8.41YY157 pKa = 10.75TSTNNLLALVNDD169 pKa = 4.48RR170 pKa = 11.84SNIYY174 pKa = 10.23DD175 pKa = 3.43KK176 pKa = 11.36QKK178 pKa = 10.76FEE180 pKa = 5.04HH181 pKa = 6.75EE182 pKa = 4.27FDD184 pKa = 3.6LKK186 pKa = 10.86IVVDD190 pKa = 4.04
MM1 pKa = 7.54AAALLEE7 pKa = 4.29AAEE10 pKa = 4.15IAAEE14 pKa = 4.18IIEE17 pKa = 4.22AAKK20 pKa = 10.33LAEE23 pKa = 4.03EE24 pKa = 5.05LGTGAYY30 pKa = 10.26DD31 pKa = 4.17LATSAYY37 pKa = 9.27RR38 pKa = 11.84GLKK41 pKa = 10.15RR42 pKa = 11.84IFGTVTQRR50 pKa = 11.84YY51 pKa = 8.38EE52 pKa = 3.67PVTNFRR58 pKa = 11.84FARR61 pKa = 11.84SFTFIEE67 pKa = 4.34LPRR70 pKa = 11.84LQRR73 pKa = 11.84VLLRR77 pKa = 11.84MQNINYY83 pKa = 9.02SLYY86 pKa = 10.49YY87 pKa = 8.16EE88 pKa = 4.17RR89 pKa = 11.84EE90 pKa = 4.04YY91 pKa = 11.51ARR93 pKa = 11.84SLVSSITHH101 pKa = 5.42DD102 pKa = 2.77HH103 pKa = 6.17SMNFFVDD110 pKa = 5.08GIFRR114 pKa = 11.84EE115 pKa = 4.3DD116 pKa = 3.48VVVVIISRR124 pKa = 11.84QPFLIPFSLLSRR136 pKa = 11.84SLSLSQHH143 pKa = 5.3PTNDD147 pKa = 2.83EE148 pKa = 4.11LTTHH152 pKa = 6.94RR153 pKa = 11.84EE154 pKa = 3.71LYY156 pKa = 8.41YY157 pKa = 10.75TSTNNLLALVNDD169 pKa = 4.48RR170 pKa = 11.84SNIYY174 pKa = 10.23DD175 pKa = 3.43KK176 pKa = 11.36QKK178 pKa = 10.76FEE180 pKa = 5.04HH181 pKa = 6.75EE182 pKa = 4.27FDD184 pKa = 3.6LKK186 pKa = 10.86IVVDD190 pKa = 4.04
Molecular weight: 21.94 kDa
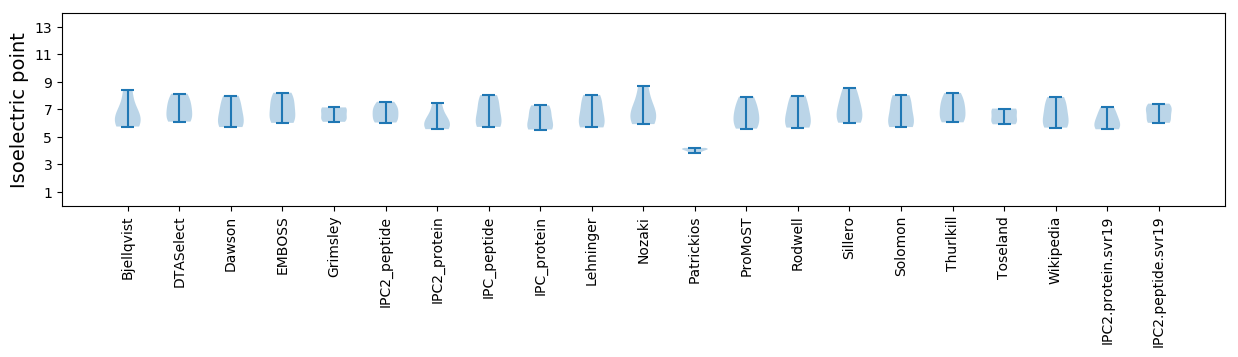
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L6YQ03|A0A1L6YQ03_9VIRU ORF2 OS=Adelphocoris suturalis virus OX=1930920 PE=4 SV=1
MM1 pKa = 7.33FCLMMLMAGIIIHH14 pKa = 6.83KK15 pKa = 9.62SMGMRR20 pKa = 11.84SMSLPAIVHH29 pKa = 5.47ATEE32 pKa = 3.87STPAVVYY39 pKa = 10.87SSDD42 pKa = 3.56SDD44 pKa = 3.95SVSIITDD51 pKa = 3.65DD52 pKa = 4.39DD53 pKa = 4.05LVDD56 pKa = 3.51YY57 pKa = 11.11SVVQRR62 pKa = 11.84RR63 pKa = 11.84SRR65 pKa = 11.84RR66 pKa = 11.84APPPEE71 pKa = 4.27RR72 pKa = 11.84PCSGSYY78 pKa = 10.16YY79 pKa = 10.88DD80 pKa = 3.87NGTWKK85 pKa = 8.89YY86 pKa = 7.89TQDD89 pKa = 3.62SCNVLADD96 pKa = 3.95RR97 pKa = 11.84CQVLNNGAPCSVKK110 pKa = 10.58CGTSSSRR117 pKa = 11.84TKK119 pKa = 10.55RR120 pKa = 11.84QTKK123 pKa = 6.17YY124 pKa = 9.08TPIVHH129 pKa = 6.35TPPRR133 pKa = 11.84QPVVTHH139 pKa = 6.33KK140 pKa = 11.22GKK142 pKa = 11.07VNVSADD148 pKa = 3.04TDD150 pKa = 3.35IYY152 pKa = 11.32YY153 pKa = 10.69LANNVTHH160 pKa = 7.19RR161 pKa = 11.84VPLEE165 pKa = 4.05DD166 pKa = 3.65LHH168 pKa = 9.08IEE170 pKa = 4.24FKK172 pKa = 11.06DD173 pKa = 4.1CIVKK177 pKa = 10.54SISLPGKK184 pKa = 10.01KK185 pKa = 8.35ITVHH189 pKa = 5.38QFYY192 pKa = 11.39SMIVNEE198 pKa = 4.32VIKK201 pKa = 10.88NKK203 pKa = 10.17NYY205 pKa = 10.13KK206 pKa = 9.98HH207 pKa = 6.55LFGPLDD213 pKa = 3.97AFSHH217 pKa = 5.45SQHH220 pKa = 5.79FRR222 pKa = 11.84DD223 pKa = 3.55PAFATFFLDD232 pKa = 4.48RR233 pKa = 11.84IAMYY237 pKa = 9.33EE238 pKa = 3.77EE239 pKa = 4.53RR240 pKa = 11.84NPTFRR245 pKa = 11.84SNLRR249 pKa = 11.84DD250 pKa = 3.37VFAAEE255 pKa = 4.28IGCTIADD262 pKa = 3.91KK263 pKa = 10.91SVCSHH268 pKa = 7.07DD269 pKa = 4.07HH270 pKa = 5.78LQHH273 pKa = 6.31VIYY276 pKa = 10.65SALDD280 pKa = 3.49LLKK283 pKa = 10.53NKK285 pKa = 9.8HH286 pKa = 4.96SSKK289 pKa = 10.9VYY291 pKa = 9.37VEE293 pKa = 4.16TDD295 pKa = 4.2FGCAIVSSTTIHH307 pKa = 6.34FGKK310 pKa = 9.99SCSPTTRR317 pKa = 11.84LRR319 pKa = 11.84TSEE322 pKa = 4.17LASNLPAICEE332 pKa = 3.9IVPIPFLHH340 pKa = 6.93RR341 pKa = 11.84SHH343 pKa = 6.25ITAPEE348 pKa = 3.86FLQILGSLNATDD360 pKa = 4.2FDD362 pKa = 3.84EE363 pKa = 4.96FAFVINKK370 pKa = 9.75RR371 pKa = 11.84IRR373 pKa = 11.84AHH375 pKa = 6.19DD376 pKa = 3.39THH378 pKa = 8.39SEE380 pKa = 4.07AKK382 pKa = 9.58EE383 pKa = 3.75HH384 pKa = 6.32LTLSMKK390 pKa = 10.3KK391 pKa = 10.06KK392 pKa = 10.3FVIEE396 pKa = 4.23DD397 pKa = 3.87FFPSRR402 pKa = 11.84SNISLNEE409 pKa = 3.77VKK411 pKa = 10.52GASVLMNDD419 pKa = 3.49SDD421 pKa = 4.31CRR423 pKa = 11.84IAIYY427 pKa = 10.25SVIFHH432 pKa = 6.64RR433 pKa = 11.84EE434 pKa = 3.35KK435 pKa = 10.97HH436 pKa = 5.58LVGTLSKK443 pKa = 10.6KK444 pKa = 10.37PEE446 pKa = 4.33DD447 pKa = 3.73SVVYY451 pKa = 8.42YY452 pKa = 9.51YY453 pKa = 10.41PKK455 pKa = 10.93VNATEE460 pKa = 4.22CC461 pKa = 3.71
MM1 pKa = 7.33FCLMMLMAGIIIHH14 pKa = 6.83KK15 pKa = 9.62SMGMRR20 pKa = 11.84SMSLPAIVHH29 pKa = 5.47ATEE32 pKa = 3.87STPAVVYY39 pKa = 10.87SSDD42 pKa = 3.56SDD44 pKa = 3.95SVSIITDD51 pKa = 3.65DD52 pKa = 4.39DD53 pKa = 4.05LVDD56 pKa = 3.51YY57 pKa = 11.11SVVQRR62 pKa = 11.84RR63 pKa = 11.84SRR65 pKa = 11.84RR66 pKa = 11.84APPPEE71 pKa = 4.27RR72 pKa = 11.84PCSGSYY78 pKa = 10.16YY79 pKa = 10.88DD80 pKa = 3.87NGTWKK85 pKa = 8.89YY86 pKa = 7.89TQDD89 pKa = 3.62SCNVLADD96 pKa = 3.95RR97 pKa = 11.84CQVLNNGAPCSVKK110 pKa = 10.58CGTSSSRR117 pKa = 11.84TKK119 pKa = 10.55RR120 pKa = 11.84QTKK123 pKa = 6.17YY124 pKa = 9.08TPIVHH129 pKa = 6.35TPPRR133 pKa = 11.84QPVVTHH139 pKa = 6.33KK140 pKa = 11.22GKK142 pKa = 11.07VNVSADD148 pKa = 3.04TDD150 pKa = 3.35IYY152 pKa = 11.32YY153 pKa = 10.69LANNVTHH160 pKa = 7.19RR161 pKa = 11.84VPLEE165 pKa = 4.05DD166 pKa = 3.65LHH168 pKa = 9.08IEE170 pKa = 4.24FKK172 pKa = 11.06DD173 pKa = 4.1CIVKK177 pKa = 10.54SISLPGKK184 pKa = 10.01KK185 pKa = 8.35ITVHH189 pKa = 5.38QFYY192 pKa = 11.39SMIVNEE198 pKa = 4.32VIKK201 pKa = 10.88NKK203 pKa = 10.17NYY205 pKa = 10.13KK206 pKa = 9.98HH207 pKa = 6.55LFGPLDD213 pKa = 3.97AFSHH217 pKa = 5.45SQHH220 pKa = 5.79FRR222 pKa = 11.84DD223 pKa = 3.55PAFATFFLDD232 pKa = 4.48RR233 pKa = 11.84IAMYY237 pKa = 9.33EE238 pKa = 3.77EE239 pKa = 4.53RR240 pKa = 11.84NPTFRR245 pKa = 11.84SNLRR249 pKa = 11.84DD250 pKa = 3.37VFAAEE255 pKa = 4.28IGCTIADD262 pKa = 3.91KK263 pKa = 10.91SVCSHH268 pKa = 7.07DD269 pKa = 4.07HH270 pKa = 5.78LQHH273 pKa = 6.31VIYY276 pKa = 10.65SALDD280 pKa = 3.49LLKK283 pKa = 10.53NKK285 pKa = 9.8HH286 pKa = 4.96SSKK289 pKa = 10.9VYY291 pKa = 9.37VEE293 pKa = 4.16TDD295 pKa = 4.2FGCAIVSSTTIHH307 pKa = 6.34FGKK310 pKa = 9.99SCSPTTRR317 pKa = 11.84LRR319 pKa = 11.84TSEE322 pKa = 4.17LASNLPAICEE332 pKa = 3.9IVPIPFLHH340 pKa = 6.93RR341 pKa = 11.84SHH343 pKa = 6.25ITAPEE348 pKa = 3.86FLQILGSLNATDD360 pKa = 4.2FDD362 pKa = 3.84EE363 pKa = 4.96FAFVINKK370 pKa = 9.75RR371 pKa = 11.84IRR373 pKa = 11.84AHH375 pKa = 6.19DD376 pKa = 3.39THH378 pKa = 8.39SEE380 pKa = 4.07AKK382 pKa = 9.58EE383 pKa = 3.75HH384 pKa = 6.32LTLSMKK390 pKa = 10.3KK391 pKa = 10.06KK392 pKa = 10.3FVIEE396 pKa = 4.23DD397 pKa = 3.87FFPSRR402 pKa = 11.84SNISLNEE409 pKa = 3.77VKK411 pKa = 10.52GASVLMNDD419 pKa = 3.49SDD421 pKa = 4.31CRR423 pKa = 11.84IAIYY427 pKa = 10.25SVIFHH432 pKa = 6.64RR433 pKa = 11.84EE434 pKa = 3.35KK435 pKa = 10.97HH436 pKa = 5.58LVGTLSKK443 pKa = 10.6KK444 pKa = 10.37PEE446 pKa = 4.33DD447 pKa = 3.73SVVYY451 pKa = 8.42YY452 pKa = 9.51YY453 pKa = 10.41PKK455 pKa = 10.93VNATEE460 pKa = 4.22CC461 pKa = 3.71
Molecular weight: 51.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3505 |
103 |
2592 |
701.0 |
79.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.191 ± 0.537 | 1.769 ± 0.509 |
6.391 ± 0.485 | 5.307 ± 0.613 |
5.05 ± 0.239 | 4.051 ± 0.466 |
3.252 ± 0.49 | 5.991 ± 0.345 |
5.364 ± 0.705 | 9.757 ± 1.333 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.854 ± 0.175 | 4.251 ± 0.379 |
5.193 ± 0.969 | 2.596 ± 0.324 |
5.735 ± 0.416 | 9.016 ± 0.631 |
5.592 ± 0.597 | 7.332 ± 0.278 |
0.656 ± 0.198 | 4.65 ± 0.327 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
