
Seal anellovirus 3
Taxonomy: Viruses; Anelloviridae; Lambdatorquevirus; Torque teno pinniped virus 3
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
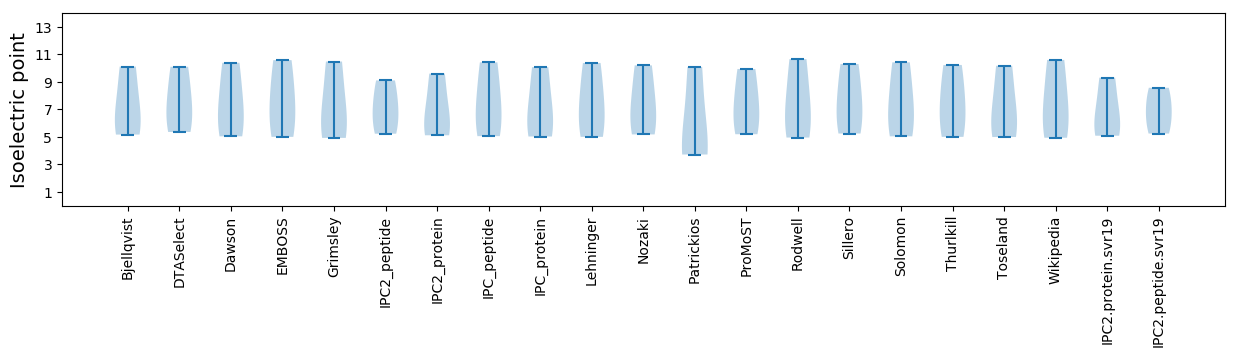
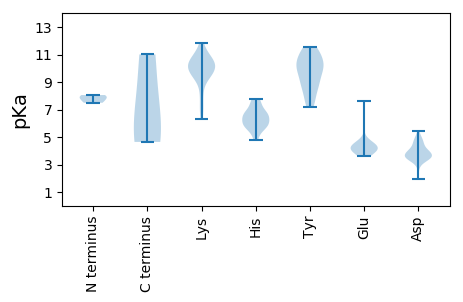
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V5NF40|V5NF40_9VIRU Capsid protein OS=Seal anellovirus 3 OX=1427156 PE=3 SV=1
MM1 pKa = 7.49MINHH5 pKa = 6.08QPHH8 pKa = 7.59AIQTTGDD15 pKa = 3.51PFYY18 pKa = 10.27HH19 pKa = 5.94QKK21 pKa = 11.18GLVLVSQSNMTQFLFGFAIASNLRR45 pKa = 11.84SLEE48 pKa = 4.03TLSTQEE54 pKa = 4.25SPAGQQRR61 pKa = 11.84RR62 pKa = 11.84SSVKK66 pKa = 10.49RR67 pKa = 11.84QDD69 pKa = 3.96LSQKK73 pKa = 10.0YY74 pKa = 9.18RR75 pKa = 11.84LAPYY79 pKa = 9.92SRR81 pKa = 11.84PDD83 pKa = 3.41LQTLGTYY90 pKa = 9.43LQEE93 pKa = 4.24TSTRR97 pKa = 11.84TASSRR102 pKa = 11.84MRR104 pKa = 11.84VLNEE108 pKa = 3.78LLDD111 pKa = 5.03LIQEE115 pKa = 4.41DD116 pKa = 4.84SQPEE120 pKa = 4.03WKK122 pKa = 9.9PIRR125 pKa = 11.84TSSSKK130 pKa = 10.73DD131 pKa = 3.19VSDD134 pKa = 4.76FDD136 pKa = 3.98TTPGSEE142 pKa = 4.36SEE144 pKa = 4.56DD145 pKa = 3.55SDD147 pKa = 4.73VSYY150 pKa = 11.4LDD152 pKa = 3.77WDD154 pKa = 3.94WEE156 pKa = 4.29DD157 pKa = 3.75EE158 pKa = 4.32PNPGGKK164 pKa = 9.88GPPLVPPTPGPKK176 pKa = 9.34AQGYY180 pKa = 7.22PPHH183 pKa = 6.79FLFF186 pKa = 5.84
MM1 pKa = 7.49MINHH5 pKa = 6.08QPHH8 pKa = 7.59AIQTTGDD15 pKa = 3.51PFYY18 pKa = 10.27HH19 pKa = 5.94QKK21 pKa = 11.18GLVLVSQSNMTQFLFGFAIASNLRR45 pKa = 11.84SLEE48 pKa = 4.03TLSTQEE54 pKa = 4.25SPAGQQRR61 pKa = 11.84RR62 pKa = 11.84SSVKK66 pKa = 10.49RR67 pKa = 11.84QDD69 pKa = 3.96LSQKK73 pKa = 10.0YY74 pKa = 9.18RR75 pKa = 11.84LAPYY79 pKa = 9.92SRR81 pKa = 11.84PDD83 pKa = 3.41LQTLGTYY90 pKa = 9.43LQEE93 pKa = 4.24TSTRR97 pKa = 11.84TASSRR102 pKa = 11.84MRR104 pKa = 11.84VLNEE108 pKa = 3.78LLDD111 pKa = 5.03LIQEE115 pKa = 4.41DD116 pKa = 4.84SQPEE120 pKa = 4.03WKK122 pKa = 9.9PIRR125 pKa = 11.84TSSSKK130 pKa = 10.73DD131 pKa = 3.19VSDD134 pKa = 4.76FDD136 pKa = 3.98TTPGSEE142 pKa = 4.36SEE144 pKa = 4.56DD145 pKa = 3.55SDD147 pKa = 4.73VSYY150 pKa = 11.4LDD152 pKa = 3.77WDD154 pKa = 3.94WEE156 pKa = 4.29DD157 pKa = 3.75EE158 pKa = 4.32PNPGGKK164 pKa = 9.88GPPLVPPTPGPKK176 pKa = 9.34AQGYY180 pKa = 7.22PPHH183 pKa = 6.79FLFF186 pKa = 5.84
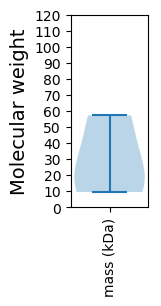
Molecular weight: 20.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5NGP8|V5NGP8_9VIRU ORF2 OS=Seal anellovirus 3 OX=1427156 PE=4 SV=1
MM1 pKa = 8.06AYY3 pKa = 9.86RR4 pKa = 11.84HH5 pKa = 5.89RR6 pKa = 11.84RR7 pKa = 11.84PFRR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 7.7RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.37RR17 pKa = 11.84RR18 pKa = 11.84WRR20 pKa = 11.84GRR22 pKa = 11.84KK23 pKa = 6.34VHH25 pKa = 6.59HH26 pKa = 6.49RR27 pKa = 11.84RR28 pKa = 11.84TWRR31 pKa = 11.84PFHH34 pKa = 6.47RR35 pKa = 11.84FGWRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84WRR43 pKa = 11.84HH44 pKa = 4.77PRR46 pKa = 11.84RR47 pKa = 11.84GSAVVRR53 pKa = 11.84ITRR56 pKa = 11.84PKK58 pKa = 7.13KK59 pKa = 9.24HH60 pKa = 6.56KK61 pKa = 10.35YY62 pKa = 7.45ITVTGWEE69 pKa = 4.22PLGNCCVTDD78 pKa = 3.65SAGAEE83 pKa = 3.85AKK85 pKa = 10.23PYY87 pKa = 10.74EE88 pKa = 4.77DD89 pKa = 4.46LDD91 pKa = 4.63KK92 pKa = 11.43DD93 pKa = 3.82VATWCTQNNNAVWHH107 pKa = 5.67GTFGHH112 pKa = 7.13HH113 pKa = 5.93YY114 pKa = 8.2LTWGLLVQRR123 pKa = 11.84AQYY126 pKa = 8.59YY127 pKa = 8.59FCKK130 pKa = 10.42FSSDD134 pKa = 3.45YY135 pKa = 10.79RR136 pKa = 11.84GYY138 pKa = 11.09DD139 pKa = 3.35YY140 pKa = 11.57LSFCGGYY147 pKa = 8.81IWLPYY152 pKa = 10.29LYY154 pKa = 10.0APYY157 pKa = 9.8MFWLDD162 pKa = 3.38PCIIDD167 pKa = 3.96PQEE170 pKa = 4.26KK171 pKa = 10.39LMNFQLYY178 pKa = 10.21KK179 pKa = 10.23KK180 pKa = 9.74DD181 pKa = 3.61SSWFHH186 pKa = 6.48PGIFMNRR193 pKa = 11.84KK194 pKa = 7.12GARR197 pKa = 11.84FIPPSYY203 pKa = 9.1QTYY206 pKa = 7.55THH208 pKa = 6.82HH209 pKa = 6.66LYY211 pKa = 10.53KK212 pKa = 10.45RR213 pKa = 11.84LRR215 pKa = 11.84VPRR218 pKa = 11.84PPTWEE223 pKa = 3.83GVYY226 pKa = 9.12TIPAATQYY234 pKa = 11.32ILTHH238 pKa = 5.73WAWTTVTTNTAFFDD252 pKa = 4.51GACNQSGTTNTCEE265 pKa = 3.61AVPWFMKK272 pKa = 9.86GHH274 pKa = 5.23QPAGKK279 pKa = 9.33FAQKK283 pKa = 8.29LTKK286 pKa = 9.56WKK288 pKa = 9.25TDD290 pKa = 3.26TADD293 pKa = 3.15GTKK296 pKa = 10.66NLDD299 pKa = 3.5CFGDD303 pKa = 3.77PRR305 pKa = 11.84SWWVNRR311 pKa = 11.84RR312 pKa = 11.84KK313 pKa = 10.2YY314 pKa = 10.56DD315 pKa = 3.82DD316 pKa = 4.44KK317 pKa = 11.83SSTTCNTDD325 pKa = 1.96NWGPFLPSKK334 pKa = 10.18GTGTGVTKK342 pKa = 10.54QYY344 pKa = 11.39DD345 pKa = 3.89SISFWFRR352 pKa = 11.84YY353 pKa = 8.89RR354 pKa = 11.84FKK356 pKa = 10.94FKK358 pKa = 11.06VSGDD362 pKa = 3.81SIYY365 pKa = 10.97TRR367 pKa = 11.84IPSRR371 pKa = 11.84TATEE375 pKa = 4.57VISEE379 pKa = 4.27APGPQSKK386 pKa = 9.51IQARR390 pKa = 11.84SILKK394 pKa = 8.11TRR396 pKa = 11.84PPDD399 pKa = 4.13LGDD402 pKa = 3.54ILAGDD407 pKa = 4.21LDD409 pKa = 4.34EE410 pKa = 7.64DD411 pKa = 5.42GILTDD416 pKa = 4.05EE417 pKa = 4.1SLEE420 pKa = 4.24RR421 pKa = 11.84IIRR424 pKa = 11.84SHH426 pKa = 7.78PGRR429 pKa = 11.84QPAGVEE435 pKa = 3.88ADD437 pKa = 3.57PDD439 pKa = 3.5QLKK442 pKa = 9.83QRR444 pKa = 11.84RR445 pKa = 11.84VRR447 pKa = 11.84LRR449 pKa = 11.84HH450 pKa = 5.9DD451 pKa = 3.57PGVRR455 pKa = 11.84KK456 pKa = 9.59RR457 pKa = 11.84RR458 pKa = 11.84LRR460 pKa = 11.84RR461 pKa = 11.84LLSGLGLGRR470 pKa = 11.84RR471 pKa = 11.84TQSGGEE477 pKa = 4.26GPPPSPPHH485 pKa = 5.82PWAA488 pKa = 4.64
MM1 pKa = 8.06AYY3 pKa = 9.86RR4 pKa = 11.84HH5 pKa = 5.89RR6 pKa = 11.84RR7 pKa = 11.84PFRR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 7.7RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.37RR17 pKa = 11.84RR18 pKa = 11.84WRR20 pKa = 11.84GRR22 pKa = 11.84KK23 pKa = 6.34VHH25 pKa = 6.59HH26 pKa = 6.49RR27 pKa = 11.84RR28 pKa = 11.84TWRR31 pKa = 11.84PFHH34 pKa = 6.47RR35 pKa = 11.84FGWRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84WRR43 pKa = 11.84HH44 pKa = 4.77PRR46 pKa = 11.84RR47 pKa = 11.84GSAVVRR53 pKa = 11.84ITRR56 pKa = 11.84PKK58 pKa = 7.13KK59 pKa = 9.24HH60 pKa = 6.56KK61 pKa = 10.35YY62 pKa = 7.45ITVTGWEE69 pKa = 4.22PLGNCCVTDD78 pKa = 3.65SAGAEE83 pKa = 3.85AKK85 pKa = 10.23PYY87 pKa = 10.74EE88 pKa = 4.77DD89 pKa = 4.46LDD91 pKa = 4.63KK92 pKa = 11.43DD93 pKa = 3.82VATWCTQNNNAVWHH107 pKa = 5.67GTFGHH112 pKa = 7.13HH113 pKa = 5.93YY114 pKa = 8.2LTWGLLVQRR123 pKa = 11.84AQYY126 pKa = 8.59YY127 pKa = 8.59FCKK130 pKa = 10.42FSSDD134 pKa = 3.45YY135 pKa = 10.79RR136 pKa = 11.84GYY138 pKa = 11.09DD139 pKa = 3.35YY140 pKa = 11.57LSFCGGYY147 pKa = 8.81IWLPYY152 pKa = 10.29LYY154 pKa = 10.0APYY157 pKa = 9.8MFWLDD162 pKa = 3.38PCIIDD167 pKa = 3.96PQEE170 pKa = 4.26KK171 pKa = 10.39LMNFQLYY178 pKa = 10.21KK179 pKa = 10.23KK180 pKa = 9.74DD181 pKa = 3.61SSWFHH186 pKa = 6.48PGIFMNRR193 pKa = 11.84KK194 pKa = 7.12GARR197 pKa = 11.84FIPPSYY203 pKa = 9.1QTYY206 pKa = 7.55THH208 pKa = 6.82HH209 pKa = 6.66LYY211 pKa = 10.53KK212 pKa = 10.45RR213 pKa = 11.84LRR215 pKa = 11.84VPRR218 pKa = 11.84PPTWEE223 pKa = 3.83GVYY226 pKa = 9.12TIPAATQYY234 pKa = 11.32ILTHH238 pKa = 5.73WAWTTVTTNTAFFDD252 pKa = 4.51GACNQSGTTNTCEE265 pKa = 3.61AVPWFMKK272 pKa = 9.86GHH274 pKa = 5.23QPAGKK279 pKa = 9.33FAQKK283 pKa = 8.29LTKK286 pKa = 9.56WKK288 pKa = 9.25TDD290 pKa = 3.26TADD293 pKa = 3.15GTKK296 pKa = 10.66NLDD299 pKa = 3.5CFGDD303 pKa = 3.77PRR305 pKa = 11.84SWWVNRR311 pKa = 11.84RR312 pKa = 11.84KK313 pKa = 10.2YY314 pKa = 10.56DD315 pKa = 3.82DD316 pKa = 4.44KK317 pKa = 11.83SSTTCNTDD325 pKa = 1.96NWGPFLPSKK334 pKa = 10.18GTGTGVTKK342 pKa = 10.54QYY344 pKa = 11.39DD345 pKa = 3.89SISFWFRR352 pKa = 11.84YY353 pKa = 8.89RR354 pKa = 11.84FKK356 pKa = 10.94FKK358 pKa = 11.06VSGDD362 pKa = 3.81SIYY365 pKa = 10.97TRR367 pKa = 11.84IPSRR371 pKa = 11.84TATEE375 pKa = 4.57VISEE379 pKa = 4.27APGPQSKK386 pKa = 9.51IQARR390 pKa = 11.84SILKK394 pKa = 8.11TRR396 pKa = 11.84PPDD399 pKa = 4.13LGDD402 pKa = 3.54ILAGDD407 pKa = 4.21LDD409 pKa = 4.34EE410 pKa = 7.64DD411 pKa = 5.42GILTDD416 pKa = 4.05EE417 pKa = 4.1SLEE420 pKa = 4.24RR421 pKa = 11.84IIRR424 pKa = 11.84SHH426 pKa = 7.78PGRR429 pKa = 11.84QPAGVEE435 pKa = 3.88ADD437 pKa = 3.57PDD439 pKa = 3.5QLKK442 pKa = 9.83QRR444 pKa = 11.84RR445 pKa = 11.84VRR447 pKa = 11.84LRR449 pKa = 11.84HH450 pKa = 5.9DD451 pKa = 3.57PGVRR455 pKa = 11.84KK456 pKa = 9.59RR457 pKa = 11.84RR458 pKa = 11.84LRR460 pKa = 11.84RR461 pKa = 11.84LLSGLGLGRR470 pKa = 11.84RR471 pKa = 11.84TQSGGEE477 pKa = 4.26GPPPSPPHH485 pKa = 5.82PWAA488 pKa = 4.64
Molecular weight: 57.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
757 |
83 |
488 |
252.3 |
29.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.756 ± 0.537 | 1.585 ± 0.615 |
6.077 ± 0.367 | 3.303 ± 0.805 |
4.227 ± 0.264 | 7.398 ± 0.614 |
2.906 ± 0.556 | 4.359 ± 1.414 |
5.02 ± 0.674 | 7.53 ± 1.301 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.453 ± 0.402 | 2.906 ± 0.412 |
7.926 ± 0.713 | 4.756 ± 1.346 |
8.983 ± 1.954 | 7.001 ± 1.905 |
7.53 ± 0.609 | 4.227 ± 0.644 |
3.435 ± 0.836 | 4.624 ± 0.556 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
