
Chitinibacter sp. 2T18
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Neisseriales; Chromobacteriaceae; Chitinibacter
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
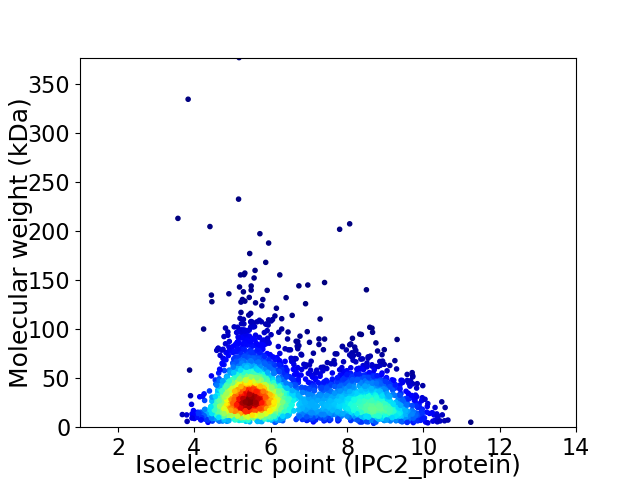
Virtual 2D-PAGE plot for 3281 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
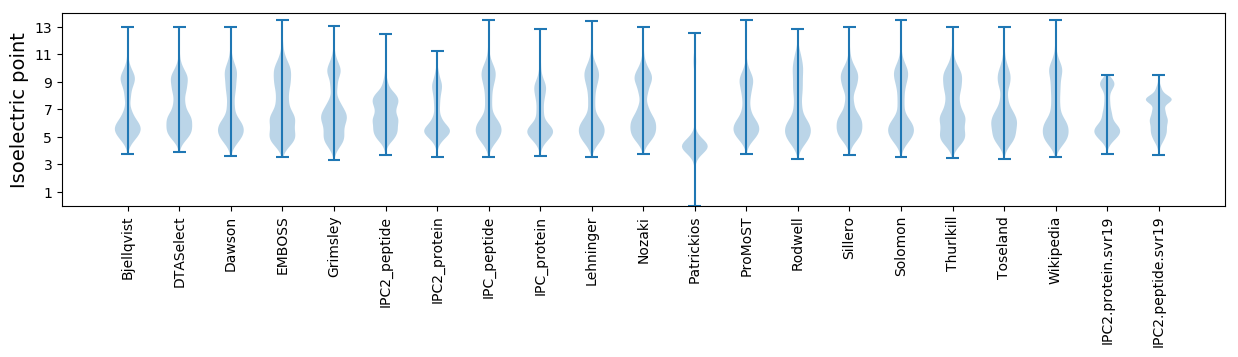
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7H9BMK3|A0A7H9BMK3_9NEIS LysE family transporter OS=Chitinibacter sp. 2T18 OX=2739434 GN=HQ393_10320 PE=4 SV=1
MM1 pKa = 7.72ASKK4 pKa = 7.71TTQTASNSAKK14 pKa = 10.16SANHH18 pKa = 6.06TGVVTQVMGEE28 pKa = 4.3VKK30 pKa = 10.54VLTLAGEE37 pKa = 4.29IKK39 pKa = 9.72TLQVGDD45 pKa = 4.05KK46 pKa = 10.73VNVGDD51 pKa = 4.74TIQTGSDD58 pKa = 3.44GAVSISLDD66 pKa = 3.3GGGTISVGRR75 pKa = 11.84SDD77 pKa = 5.6SMPLTNEE84 pKa = 3.32MFAALVAPADD94 pKa = 3.77NAAADD99 pKa = 3.56AAKK102 pKa = 9.91IQEE105 pKa = 5.07LIAQGADD112 pKa = 3.36PTQVAAASAAGAGGGEE128 pKa = 4.44DD129 pKa = 3.56GGHH132 pKa = 5.8SFVVLDD138 pKa = 3.87SPEE141 pKa = 3.86SRR143 pKa = 11.84VAVDD147 pKa = 3.09TGFATTPVIQSVSLIPPDD165 pKa = 3.61ASNAITPTNTPPVANNDD182 pKa = 3.45TFTPASPPNALNDD195 pKa = 3.94ALNTNEE201 pKa = 4.53DD202 pKa = 4.06TPLVIQGTTLLANDD216 pKa = 4.38SDD218 pKa = 5.0ADD220 pKa = 4.1GNPLTIIGVGTAGNGTVILNADD242 pKa = 3.56GSITYY247 pKa = 8.4TPNANYY253 pKa = 10.2NGPDD257 pKa = 3.15SFTYY261 pKa = 9.94TISDD265 pKa = 3.88GQGGTSTATVTVGVTPVNDD284 pKa = 3.83PPVLLNPEE292 pKa = 3.94NLAPLGDD299 pKa = 4.3DD300 pKa = 3.65VSVTTPEE307 pKa = 3.93DD308 pKa = 3.81TPVGGKK314 pKa = 9.24LAATDD319 pKa = 3.56VDD321 pKa = 4.24GDD323 pKa = 4.04TLTFSLTGNQPAHH336 pKa = 5.73GTVTVNADD344 pKa = 3.54GTWLYY349 pKa = 10.23TPNTNYY355 pKa = 10.53NGPDD359 pKa = 3.26SFTALVDD366 pKa = 4.0DD367 pKa = 5.13GKK369 pKa = 11.36GGTDD373 pKa = 3.13TLVVNIGVTPVNDD386 pKa = 3.64PPVLVNPEE394 pKa = 3.72NLAPLGDD401 pKa = 4.3DD402 pKa = 3.65VSVTTPEE409 pKa = 3.88DD410 pKa = 3.72TPIGGKK416 pKa = 9.37LAATDD421 pKa = 3.9ADD423 pKa = 4.09GDD425 pKa = 4.14TLTFSLTGNQPAHH438 pKa = 5.73GTVTVNADD446 pKa = 3.54GTWLYY451 pKa = 10.23TPNTNYY457 pKa = 10.53NGPDD461 pKa = 3.26SFTAMVDD468 pKa = 3.47DD469 pKa = 4.98GKK471 pKa = 11.36GGTDD475 pKa = 3.13TLVVNIGVTPVNDD488 pKa = 3.65PPVIVDD494 pKa = 4.16PEE496 pKa = 4.08NLAPLGDD503 pKa = 4.3DD504 pKa = 3.65VSVTTPEE511 pKa = 3.88DD512 pKa = 3.72TPIGGKK518 pKa = 9.37LAATDD523 pKa = 3.86ADD525 pKa = 4.11GDD527 pKa = 4.3SLTFSLTGNQPAHH540 pKa = 5.73GTVTVNADD548 pKa = 3.54GTWLYY553 pKa = 10.23TPNTNYY559 pKa = 10.53NGPDD563 pKa = 3.26SFTAMVDD570 pKa = 3.47DD571 pKa = 4.98GKK573 pKa = 11.36GGTDD577 pKa = 3.13TLVVNIGVTPLNAPPTITTDD597 pKa = 3.06NGNEE601 pKa = 4.18GEE603 pKa = 4.6GNDD606 pKa = 3.66TVYY609 pKa = 11.04EE610 pKa = 4.22SGLATGTNAGVSTITASGTFTVADD634 pKa = 4.0ADD636 pKa = 4.03GLGDD640 pKa = 3.37IHH642 pKa = 7.53SVTVAGTTFVIGAGGLADD660 pKa = 4.44LAALVGQTANTSHH673 pKa = 6.8GSVLIDD679 pKa = 3.45SYY681 pKa = 11.86NNGVFGYY688 pKa = 8.5TYY690 pKa = 10.31TLTSPVTSGSAEE702 pKa = 3.57SDD704 pKa = 3.57SFNVSVADD712 pKa = 4.06GSGASAPVTVTINIVDD728 pKa = 4.36DD729 pKa = 4.79APVSTSEE736 pKa = 4.21HH737 pKa = 5.71VSVTEE742 pKa = 3.95DD743 pKa = 3.0AASNVVSGNVLDD755 pKa = 4.58NDD757 pKa = 3.62VSGADD762 pKa = 3.62SPKK765 pKa = 10.84SFAGWSTGDD774 pKa = 3.39AATITALQNFGTLVQDD790 pKa = 4.68SDD792 pKa = 4.13GHH794 pKa = 5.6WSYY797 pKa = 10.13TLNNGLSATQALTGSDD813 pKa = 3.02QTDD816 pKa = 3.67YY817 pKa = 11.13VLHH820 pKa = 5.59YY821 pKa = 8.91TVQDD825 pKa = 3.43ADD827 pKa = 4.01GTTSPATLTITIKK840 pKa = 10.56GANDD844 pKa = 3.85GPTITTDD851 pKa = 2.95NGNDD855 pKa = 3.82GEE857 pKa = 4.78GNDD860 pKa = 3.81TVYY863 pKa = 11.04EE864 pKa = 4.22SGLATGTNAGVSTITASGTFTVADD888 pKa = 4.0ADD890 pKa = 4.03GLGDD894 pKa = 3.37IHH896 pKa = 7.53SVTVAGTTFVIGTGTGEE913 pKa = 4.26FVDD916 pKa = 4.87LAALVGQTANTSHH929 pKa = 6.8GSVLIDD935 pKa = 3.35SYY937 pKa = 12.14NNGVYY942 pKa = 10.19SYY944 pKa = 10.22TYY946 pKa = 8.69TLSGAVPDD954 pKa = 3.97IANQVEE960 pKa = 4.49TDD962 pKa = 3.42SFTVSVSDD970 pKa = 3.95GVAPAASATITIEE983 pKa = 3.9INDD986 pKa = 5.47DD987 pKa = 3.36IPIAKK992 pKa = 9.76ADD994 pKa = 3.96SNNISANDD1002 pKa = 3.54FNSEE1006 pKa = 4.06FEE1008 pKa = 4.56GVQEE1012 pKa = 4.21LPAVQTSGNVLLGDD1026 pKa = 4.14LNGGVADD1033 pKa = 4.05VQGADD1038 pKa = 3.4QPAKK1042 pKa = 7.84VTQVIFGTNVQAVTTDD1058 pKa = 3.26APTSIDD1064 pKa = 3.26GKK1066 pKa = 11.13YY1067 pKa = 8.8GTLLLNSDD1075 pKa = 3.55GSYY1078 pKa = 10.29TYY1080 pKa = 10.6TLNNSSNAVQEE1091 pKa = 4.41LGNNEE1096 pKa = 4.15HH1097 pKa = 6.52PTDD1100 pKa = 3.6VFSYY1104 pKa = 10.15SIKK1107 pKa = 10.94DD1108 pKa = 3.29ADD1110 pKa = 3.81GDD1112 pKa = 4.14VSNLTSLTITVNGADD1127 pKa = 3.98DD1128 pKa = 6.28AIVLDD1133 pKa = 4.17TKK1135 pKa = 11.26LSGEE1139 pKa = 4.28VSSGTVYY1146 pKa = 10.56EE1147 pKa = 4.87AGLSTGSKK1155 pKa = 10.56ADD1157 pKa = 3.46GDD1159 pKa = 4.1ATTDD1163 pKa = 3.31PTTFTSSFTIKK1174 pKa = 10.67SIDD1177 pKa = 3.7GLDD1180 pKa = 3.32HH1181 pKa = 5.29VTIGTGANTARR1192 pKa = 11.84VNFTQAPDD1200 pKa = 4.19GSWSASVADD1209 pKa = 4.24SSADD1213 pKa = 3.42SALGNSLTVTNAVYY1227 pKa = 10.74SSTSHH1232 pKa = 5.48TWTFSYY1238 pKa = 10.4SYY1240 pKa = 9.65TLNAPEE1246 pKa = 4.1THH1248 pKa = 6.41SAPGNDD1254 pKa = 3.28NVSDD1258 pKa = 4.02TLSVMALEE1266 pKa = 4.84TDD1268 pKa = 3.53GSTANGTLSITVVDD1282 pKa = 4.85DD1283 pKa = 3.81VPTAKK1288 pKa = 10.59ADD1290 pKa = 3.75FDD1292 pKa = 4.86SVTEE1296 pKa = 4.41DD1297 pKa = 3.21ASSTIATGNVITGVDD1312 pKa = 3.58LAGSHH1317 pKa = 7.34DD1318 pKa = 4.57SNSSDD1323 pKa = 3.66GVADD1327 pKa = 3.52VQGADD1332 pKa = 3.29RR1333 pKa = 11.84NVMVTHH1339 pKa = 7.01IGFGTNVQEE1348 pKa = 4.4VHH1350 pKa = 6.16TGTPTSINGQYY1361 pKa = 11.16GSLSLNSDD1369 pKa = 3.1GSYY1372 pKa = 10.09TYY1374 pKa = 10.94SLHH1377 pKa = 6.2NSSNAVQEE1385 pKa = 4.41LGNNEE1390 pKa = 4.1HH1391 pKa = 6.24PTEE1394 pKa = 4.12VFTYY1398 pKa = 9.5TITDD1402 pKa = 3.44ADD1404 pKa = 4.23GDD1406 pKa = 4.04RR1407 pKa = 11.84SSQTLTISVNGADD1420 pKa = 5.29DD1421 pKa = 6.6AIALDD1426 pKa = 3.67NKK1428 pKa = 10.7LITEE1432 pKa = 4.5TNSGTVNEE1440 pKa = 4.82AGLPSGSLADD1450 pKa = 4.37SDD1452 pKa = 4.33PTTQPTSLAGSFTVKK1467 pKa = 10.61SIDD1470 pKa = 3.67GLDD1473 pKa = 3.31HH1474 pKa = 5.29VTIGTGAGNTATVSFTQAADD1494 pKa = 4.23GSWTPHH1500 pKa = 5.22FTDD1503 pKa = 4.94PSAVSALGNSLSITGSSYY1521 pKa = 11.18NAADD1525 pKa = 3.68HH1526 pKa = 6.25TWTLNYY1532 pKa = 10.16TYY1534 pKa = 9.87TLQAAEE1540 pKa = 4.16THH1542 pKa = 5.97SAAGNDD1548 pKa = 3.8TLIDD1552 pKa = 3.78TLAVSAIEE1560 pKa = 4.31TDD1562 pKa = 3.24GSTANGTIRR1571 pKa = 11.84IGVVDD1576 pKa = 5.03DD1577 pKa = 4.51APIAKK1582 pKa = 9.98DD1583 pKa = 3.19ISLSSATHH1591 pKa = 4.51VVEE1594 pKa = 4.39TNLVLTIDD1602 pKa = 4.09VSGSMQGSKK1611 pKa = 10.79LDD1613 pKa = 3.84LAKK1616 pKa = 10.44QALSNVIDD1624 pKa = 3.83QYY1626 pKa = 11.94DD1627 pKa = 3.33ALGDD1631 pKa = 3.66VRR1633 pKa = 11.84IMLVNFSSDD1642 pKa = 2.89ASLQGSNTWLSIGAAKK1658 pKa = 10.54ALINLLSAGGNTNFDD1673 pKa = 3.42AALAQITTHH1682 pKa = 5.98YY1683 pKa = 10.64NDD1685 pKa = 3.33YY1686 pKa = 11.38GKK1688 pKa = 8.8LTGEE1692 pKa = 4.5GVQNVSYY1699 pKa = 9.95FLSDD1703 pKa = 3.49GVPTVGGGITGSEE1716 pKa = 3.66IGAWTHH1722 pKa = 6.31FVDD1725 pKa = 4.72INDD1728 pKa = 3.64IKK1730 pKa = 11.27SLAFGIGTGATLSTLEE1746 pKa = 4.54PIAYY1750 pKa = 9.99DD1751 pKa = 3.51GVTNTDD1757 pKa = 3.21PSSGVTLVTNASNLPSALVNSAQPVLNFSLLDD1789 pKa = 3.56NGGSFGADD1797 pKa = 2.42GGHH1800 pKa = 6.2VNNIIIDD1807 pKa = 3.53GHH1809 pKa = 5.56TYY1811 pKa = 10.97SFGDD1815 pKa = 3.38ASKK1818 pKa = 9.99TMTQTGNIYY1827 pKa = 10.47SYY1829 pKa = 11.5DD1830 pKa = 3.63ATTHH1834 pKa = 6.0EE1835 pKa = 4.77LTVNTQTGSTLNGGTFTVDD1854 pKa = 2.46MDD1856 pKa = 3.61TGVAHH1861 pKa = 6.01FTGPASLTTALTEE1874 pKa = 4.44NISFQLIDD1882 pKa = 3.47NDD1884 pKa = 4.06GDD1886 pKa = 3.69VSASKK1891 pKa = 10.93NLGVTIAAADD1901 pKa = 3.78SGSNVYY1907 pKa = 8.81DD1908 pKa = 3.38TTGGHH1913 pKa = 6.08TLTGTAFNDD1922 pKa = 4.38TIDD1925 pKa = 4.41GNDD1928 pKa = 3.68GNDD1931 pKa = 4.02TINGNAGHH1939 pKa = 7.14DD1940 pKa = 3.8RR1941 pKa = 11.84LDD1943 pKa = 3.83GGTGNDD1949 pKa = 3.83TLNGGDD1955 pKa = 4.38GNDD1958 pKa = 3.32VLYY1961 pKa = 11.12GGIGDD1966 pKa = 3.94DD1967 pKa = 4.69TINGDD1972 pKa = 3.48SGNDD1976 pKa = 3.29VLIGGKK1982 pKa = 10.59GNDD1985 pKa = 3.5ILNGGSGVDD1994 pKa = 3.14TFVFSSTASDD2004 pKa = 3.34NGKK2007 pKa = 8.69DD2008 pKa = 3.58TIQDD2012 pKa = 3.17FTVAPKK2018 pKa = 10.62VAGLVQGDD2026 pKa = 3.72ILDD2029 pKa = 3.93ISDD2032 pKa = 5.06LLTPSTAATFNSSEE2046 pKa = 3.98AAAGAFLQFVQDD2058 pKa = 3.83GANTKK2063 pKa = 10.34VMFDD2067 pKa = 3.62ADD2069 pKa = 3.75GSGTSAAPVQVATLTGVNATDD2090 pKa = 4.42LLHH2093 pKa = 6.93HH2094 pKa = 6.77LLDD2097 pKa = 3.92NGEE2100 pKa = 4.36IKK2102 pKa = 9.91TSHH2105 pKa = 6.21
MM1 pKa = 7.72ASKK4 pKa = 7.71TTQTASNSAKK14 pKa = 10.16SANHH18 pKa = 6.06TGVVTQVMGEE28 pKa = 4.3VKK30 pKa = 10.54VLTLAGEE37 pKa = 4.29IKK39 pKa = 9.72TLQVGDD45 pKa = 4.05KK46 pKa = 10.73VNVGDD51 pKa = 4.74TIQTGSDD58 pKa = 3.44GAVSISLDD66 pKa = 3.3GGGTISVGRR75 pKa = 11.84SDD77 pKa = 5.6SMPLTNEE84 pKa = 3.32MFAALVAPADD94 pKa = 3.77NAAADD99 pKa = 3.56AAKK102 pKa = 9.91IQEE105 pKa = 5.07LIAQGADD112 pKa = 3.36PTQVAAASAAGAGGGEE128 pKa = 4.44DD129 pKa = 3.56GGHH132 pKa = 5.8SFVVLDD138 pKa = 3.87SPEE141 pKa = 3.86SRR143 pKa = 11.84VAVDD147 pKa = 3.09TGFATTPVIQSVSLIPPDD165 pKa = 3.61ASNAITPTNTPPVANNDD182 pKa = 3.45TFTPASPPNALNDD195 pKa = 3.94ALNTNEE201 pKa = 4.53DD202 pKa = 4.06TPLVIQGTTLLANDD216 pKa = 4.38SDD218 pKa = 5.0ADD220 pKa = 4.1GNPLTIIGVGTAGNGTVILNADD242 pKa = 3.56GSITYY247 pKa = 8.4TPNANYY253 pKa = 10.2NGPDD257 pKa = 3.15SFTYY261 pKa = 9.94TISDD265 pKa = 3.88GQGGTSTATVTVGVTPVNDD284 pKa = 3.83PPVLLNPEE292 pKa = 3.94NLAPLGDD299 pKa = 4.3DD300 pKa = 3.65VSVTTPEE307 pKa = 3.93DD308 pKa = 3.81TPVGGKK314 pKa = 9.24LAATDD319 pKa = 3.56VDD321 pKa = 4.24GDD323 pKa = 4.04TLTFSLTGNQPAHH336 pKa = 5.73GTVTVNADD344 pKa = 3.54GTWLYY349 pKa = 10.23TPNTNYY355 pKa = 10.53NGPDD359 pKa = 3.26SFTALVDD366 pKa = 4.0DD367 pKa = 5.13GKK369 pKa = 11.36GGTDD373 pKa = 3.13TLVVNIGVTPVNDD386 pKa = 3.64PPVLVNPEE394 pKa = 3.72NLAPLGDD401 pKa = 4.3DD402 pKa = 3.65VSVTTPEE409 pKa = 3.88DD410 pKa = 3.72TPIGGKK416 pKa = 9.37LAATDD421 pKa = 3.9ADD423 pKa = 4.09GDD425 pKa = 4.14TLTFSLTGNQPAHH438 pKa = 5.73GTVTVNADD446 pKa = 3.54GTWLYY451 pKa = 10.23TPNTNYY457 pKa = 10.53NGPDD461 pKa = 3.26SFTAMVDD468 pKa = 3.47DD469 pKa = 4.98GKK471 pKa = 11.36GGTDD475 pKa = 3.13TLVVNIGVTPVNDD488 pKa = 3.65PPVIVDD494 pKa = 4.16PEE496 pKa = 4.08NLAPLGDD503 pKa = 4.3DD504 pKa = 3.65VSVTTPEE511 pKa = 3.88DD512 pKa = 3.72TPIGGKK518 pKa = 9.37LAATDD523 pKa = 3.86ADD525 pKa = 4.11GDD527 pKa = 4.3SLTFSLTGNQPAHH540 pKa = 5.73GTVTVNADD548 pKa = 3.54GTWLYY553 pKa = 10.23TPNTNYY559 pKa = 10.53NGPDD563 pKa = 3.26SFTAMVDD570 pKa = 3.47DD571 pKa = 4.98GKK573 pKa = 11.36GGTDD577 pKa = 3.13TLVVNIGVTPLNAPPTITTDD597 pKa = 3.06NGNEE601 pKa = 4.18GEE603 pKa = 4.6GNDD606 pKa = 3.66TVYY609 pKa = 11.04EE610 pKa = 4.22SGLATGTNAGVSTITASGTFTVADD634 pKa = 4.0ADD636 pKa = 4.03GLGDD640 pKa = 3.37IHH642 pKa = 7.53SVTVAGTTFVIGAGGLADD660 pKa = 4.44LAALVGQTANTSHH673 pKa = 6.8GSVLIDD679 pKa = 3.45SYY681 pKa = 11.86NNGVFGYY688 pKa = 8.5TYY690 pKa = 10.31TLTSPVTSGSAEE702 pKa = 3.57SDD704 pKa = 3.57SFNVSVADD712 pKa = 4.06GSGASAPVTVTINIVDD728 pKa = 4.36DD729 pKa = 4.79APVSTSEE736 pKa = 4.21HH737 pKa = 5.71VSVTEE742 pKa = 3.95DD743 pKa = 3.0AASNVVSGNVLDD755 pKa = 4.58NDD757 pKa = 3.62VSGADD762 pKa = 3.62SPKK765 pKa = 10.84SFAGWSTGDD774 pKa = 3.39AATITALQNFGTLVQDD790 pKa = 4.68SDD792 pKa = 4.13GHH794 pKa = 5.6WSYY797 pKa = 10.13TLNNGLSATQALTGSDD813 pKa = 3.02QTDD816 pKa = 3.67YY817 pKa = 11.13VLHH820 pKa = 5.59YY821 pKa = 8.91TVQDD825 pKa = 3.43ADD827 pKa = 4.01GTTSPATLTITIKK840 pKa = 10.56GANDD844 pKa = 3.85GPTITTDD851 pKa = 2.95NGNDD855 pKa = 3.82GEE857 pKa = 4.78GNDD860 pKa = 3.81TVYY863 pKa = 11.04EE864 pKa = 4.22SGLATGTNAGVSTITASGTFTVADD888 pKa = 4.0ADD890 pKa = 4.03GLGDD894 pKa = 3.37IHH896 pKa = 7.53SVTVAGTTFVIGTGTGEE913 pKa = 4.26FVDD916 pKa = 4.87LAALVGQTANTSHH929 pKa = 6.8GSVLIDD935 pKa = 3.35SYY937 pKa = 12.14NNGVYY942 pKa = 10.19SYY944 pKa = 10.22TYY946 pKa = 8.69TLSGAVPDD954 pKa = 3.97IANQVEE960 pKa = 4.49TDD962 pKa = 3.42SFTVSVSDD970 pKa = 3.95GVAPAASATITIEE983 pKa = 3.9INDD986 pKa = 5.47DD987 pKa = 3.36IPIAKK992 pKa = 9.76ADD994 pKa = 3.96SNNISANDD1002 pKa = 3.54FNSEE1006 pKa = 4.06FEE1008 pKa = 4.56GVQEE1012 pKa = 4.21LPAVQTSGNVLLGDD1026 pKa = 4.14LNGGVADD1033 pKa = 4.05VQGADD1038 pKa = 3.4QPAKK1042 pKa = 7.84VTQVIFGTNVQAVTTDD1058 pKa = 3.26APTSIDD1064 pKa = 3.26GKK1066 pKa = 11.13YY1067 pKa = 8.8GTLLLNSDD1075 pKa = 3.55GSYY1078 pKa = 10.29TYY1080 pKa = 10.6TLNNSSNAVQEE1091 pKa = 4.41LGNNEE1096 pKa = 4.15HH1097 pKa = 6.52PTDD1100 pKa = 3.6VFSYY1104 pKa = 10.15SIKK1107 pKa = 10.94DD1108 pKa = 3.29ADD1110 pKa = 3.81GDD1112 pKa = 4.14VSNLTSLTITVNGADD1127 pKa = 3.98DD1128 pKa = 6.28AIVLDD1133 pKa = 4.17TKK1135 pKa = 11.26LSGEE1139 pKa = 4.28VSSGTVYY1146 pKa = 10.56EE1147 pKa = 4.87AGLSTGSKK1155 pKa = 10.56ADD1157 pKa = 3.46GDD1159 pKa = 4.1ATTDD1163 pKa = 3.31PTTFTSSFTIKK1174 pKa = 10.67SIDD1177 pKa = 3.7GLDD1180 pKa = 3.32HH1181 pKa = 5.29VTIGTGANTARR1192 pKa = 11.84VNFTQAPDD1200 pKa = 4.19GSWSASVADD1209 pKa = 4.24SSADD1213 pKa = 3.42SALGNSLTVTNAVYY1227 pKa = 10.74SSTSHH1232 pKa = 5.48TWTFSYY1238 pKa = 10.4SYY1240 pKa = 9.65TLNAPEE1246 pKa = 4.1THH1248 pKa = 6.41SAPGNDD1254 pKa = 3.28NVSDD1258 pKa = 4.02TLSVMALEE1266 pKa = 4.84TDD1268 pKa = 3.53GSTANGTLSITVVDD1282 pKa = 4.85DD1283 pKa = 3.81VPTAKK1288 pKa = 10.59ADD1290 pKa = 3.75FDD1292 pKa = 4.86SVTEE1296 pKa = 4.41DD1297 pKa = 3.21ASSTIATGNVITGVDD1312 pKa = 3.58LAGSHH1317 pKa = 7.34DD1318 pKa = 4.57SNSSDD1323 pKa = 3.66GVADD1327 pKa = 3.52VQGADD1332 pKa = 3.29RR1333 pKa = 11.84NVMVTHH1339 pKa = 7.01IGFGTNVQEE1348 pKa = 4.4VHH1350 pKa = 6.16TGTPTSINGQYY1361 pKa = 11.16GSLSLNSDD1369 pKa = 3.1GSYY1372 pKa = 10.09TYY1374 pKa = 10.94SLHH1377 pKa = 6.2NSSNAVQEE1385 pKa = 4.41LGNNEE1390 pKa = 4.1HH1391 pKa = 6.24PTEE1394 pKa = 4.12VFTYY1398 pKa = 9.5TITDD1402 pKa = 3.44ADD1404 pKa = 4.23GDD1406 pKa = 4.04RR1407 pKa = 11.84SSQTLTISVNGADD1420 pKa = 5.29DD1421 pKa = 6.6AIALDD1426 pKa = 3.67NKK1428 pKa = 10.7LITEE1432 pKa = 4.5TNSGTVNEE1440 pKa = 4.82AGLPSGSLADD1450 pKa = 4.37SDD1452 pKa = 4.33PTTQPTSLAGSFTVKK1467 pKa = 10.61SIDD1470 pKa = 3.67GLDD1473 pKa = 3.31HH1474 pKa = 5.29VTIGTGAGNTATVSFTQAADD1494 pKa = 4.23GSWTPHH1500 pKa = 5.22FTDD1503 pKa = 4.94PSAVSALGNSLSITGSSYY1521 pKa = 11.18NAADD1525 pKa = 3.68HH1526 pKa = 6.25TWTLNYY1532 pKa = 10.16TYY1534 pKa = 9.87TLQAAEE1540 pKa = 4.16THH1542 pKa = 5.97SAAGNDD1548 pKa = 3.8TLIDD1552 pKa = 3.78TLAVSAIEE1560 pKa = 4.31TDD1562 pKa = 3.24GSTANGTIRR1571 pKa = 11.84IGVVDD1576 pKa = 5.03DD1577 pKa = 4.51APIAKK1582 pKa = 9.98DD1583 pKa = 3.19ISLSSATHH1591 pKa = 4.51VVEE1594 pKa = 4.39TNLVLTIDD1602 pKa = 4.09VSGSMQGSKK1611 pKa = 10.79LDD1613 pKa = 3.84LAKK1616 pKa = 10.44QALSNVIDD1624 pKa = 3.83QYY1626 pKa = 11.94DD1627 pKa = 3.33ALGDD1631 pKa = 3.66VRR1633 pKa = 11.84IMLVNFSSDD1642 pKa = 2.89ASLQGSNTWLSIGAAKK1658 pKa = 10.54ALINLLSAGGNTNFDD1673 pKa = 3.42AALAQITTHH1682 pKa = 5.98YY1683 pKa = 10.64NDD1685 pKa = 3.33YY1686 pKa = 11.38GKK1688 pKa = 8.8LTGEE1692 pKa = 4.5GVQNVSYY1699 pKa = 9.95FLSDD1703 pKa = 3.49GVPTVGGGITGSEE1716 pKa = 3.66IGAWTHH1722 pKa = 6.31FVDD1725 pKa = 4.72INDD1728 pKa = 3.64IKK1730 pKa = 11.27SLAFGIGTGATLSTLEE1746 pKa = 4.54PIAYY1750 pKa = 9.99DD1751 pKa = 3.51GVTNTDD1757 pKa = 3.21PSSGVTLVTNASNLPSALVNSAQPVLNFSLLDD1789 pKa = 3.56NGGSFGADD1797 pKa = 2.42GGHH1800 pKa = 6.2VNNIIIDD1807 pKa = 3.53GHH1809 pKa = 5.56TYY1811 pKa = 10.97SFGDD1815 pKa = 3.38ASKK1818 pKa = 9.99TMTQTGNIYY1827 pKa = 10.47SYY1829 pKa = 11.5DD1830 pKa = 3.63ATTHH1834 pKa = 6.0EE1835 pKa = 4.77LTVNTQTGSTLNGGTFTVDD1854 pKa = 2.46MDD1856 pKa = 3.61TGVAHH1861 pKa = 6.01FTGPASLTTALTEE1874 pKa = 4.44NISFQLIDD1882 pKa = 3.47NDD1884 pKa = 4.06GDD1886 pKa = 3.69VSASKK1891 pKa = 10.93NLGVTIAAADD1901 pKa = 3.78SGSNVYY1907 pKa = 8.81DD1908 pKa = 3.38TTGGHH1913 pKa = 6.08TLTGTAFNDD1922 pKa = 4.38TIDD1925 pKa = 4.41GNDD1928 pKa = 3.68GNDD1931 pKa = 4.02TINGNAGHH1939 pKa = 7.14DD1940 pKa = 3.8RR1941 pKa = 11.84LDD1943 pKa = 3.83GGTGNDD1949 pKa = 3.83TLNGGDD1955 pKa = 4.38GNDD1958 pKa = 3.32VLYY1961 pKa = 11.12GGIGDD1966 pKa = 3.94DD1967 pKa = 4.69TINGDD1972 pKa = 3.48SGNDD1976 pKa = 3.29VLIGGKK1982 pKa = 10.59GNDD1985 pKa = 3.5ILNGGSGVDD1994 pKa = 3.14TFVFSSTASDD2004 pKa = 3.34NGKK2007 pKa = 8.69DD2008 pKa = 3.58TIQDD2012 pKa = 3.17FTVAPKK2018 pKa = 10.62VAGLVQGDD2026 pKa = 3.72ILDD2029 pKa = 3.93ISDD2032 pKa = 5.06LLTPSTAATFNSSEE2046 pKa = 3.98AAAGAFLQFVQDD2058 pKa = 3.83GANTKK2063 pKa = 10.34VMFDD2067 pKa = 3.62ADD2069 pKa = 3.75GSGTSAAPVQVATLTGVNATDD2090 pKa = 4.42LLHH2093 pKa = 6.93HH2094 pKa = 6.77LLDD2097 pKa = 3.92NGEE2100 pKa = 4.36IKK2102 pKa = 9.91TSHH2105 pKa = 6.21
Molecular weight: 212.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7H9BLL4|A0A7H9BLL4_9NEIS HD-GYP domain-containing protein OS=Chitinibacter sp. 2T18 OX=2739434 GN=HQ393_13515 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.58RR14 pKa = 11.84THH16 pKa = 6.03GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSVGGVRR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.58RR14 pKa = 11.84THH16 pKa = 6.03GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSVGGVRR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1045249 |
37 |
3629 |
318.6 |
35.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.024 ± 0.053 | 0.974 ± 0.013 |
5.184 ± 0.032 | 5.61 ± 0.043 |
3.712 ± 0.03 | 7.143 ± 0.042 |
2.241 ± 0.023 | 5.785 ± 0.038 |
4.556 ± 0.039 | 10.972 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.559 ± 0.024 | 3.727 ± 0.036 |
4.403 ± 0.03 | 4.865 ± 0.04 |
5.261 ± 0.047 | 5.954 ± 0.04 |
5.162 ± 0.055 | 6.794 ± 0.041 |
1.413 ± 0.017 | 2.66 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
