
Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) (Yeast) (Candida sphaerica)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Kluyveromyces; Kluyveromyces lactis
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
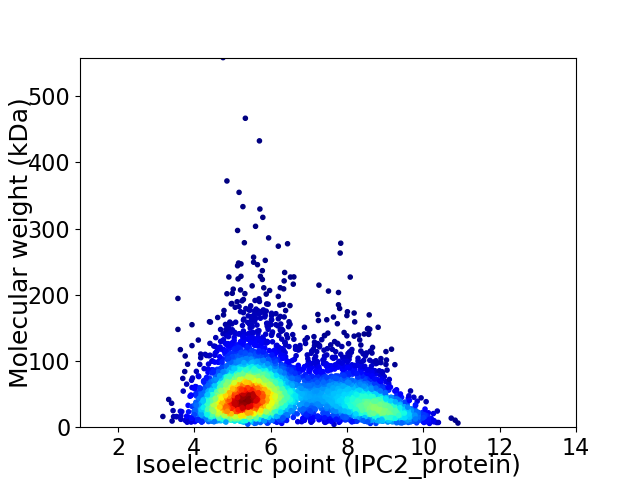
Virtual 2D-PAGE plot for 5071 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
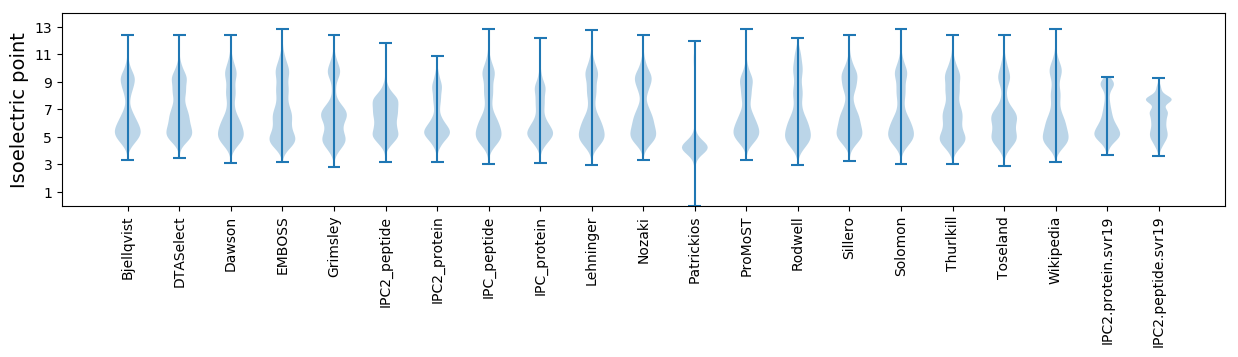
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q6CVA8|HSE1_KLULA Class E vacuolar protein-sorting machinery protein HSE1 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=HSE1 PE=3 SV=1
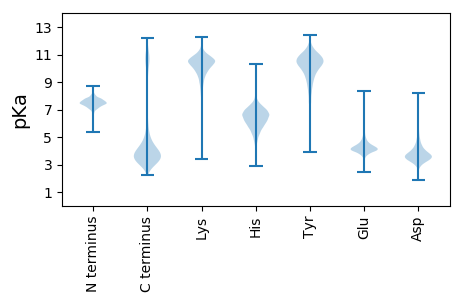
MM1 pKa = 6.79KK2 pKa = 9.98TSVVNVTILLACGVPLASALRR23 pKa = 11.84TVTSDD28 pKa = 3.22YY29 pKa = 11.59DD30 pKa = 4.15VISDD34 pKa = 3.77TVLDD38 pKa = 3.55VGGAIFNGNVQIEE51 pKa = 4.17EE52 pKa = 4.36DD53 pKa = 3.55KK54 pKa = 11.2FLALVSGTTEE64 pKa = 3.95YY65 pKa = 10.85FNKK68 pKa = 10.55DD69 pKa = 3.97LIVDD73 pKa = 4.17GALYY77 pKa = 10.13IGSKK81 pKa = 9.63IQTLGLDD88 pKa = 3.69VKK90 pKa = 11.28VLGSTQKK97 pKa = 10.02IQNTGTIVLNGHH109 pKa = 5.72NTTLPSTYY117 pKa = 9.46YY118 pKa = 9.79WVGEE122 pKa = 4.1SFKK125 pKa = 11.19NDD127 pKa = 3.02GEE129 pKa = 4.57IYY131 pKa = 10.14WIAQSSLIGSLYY143 pKa = 10.35EE144 pKa = 4.54IIPSSSFEE152 pKa = 4.4NNGLLSFTHH161 pKa = 5.9EE162 pKa = 4.22NGRR165 pKa = 11.84KK166 pKa = 9.93GGTLTLGNLGSAITNSGTICLKK188 pKa = 10.43GVVYY192 pKa = 9.93KK193 pKa = 10.31QSGMVSGEE201 pKa = 4.2SGCISIGDD209 pKa = 3.89EE210 pKa = 4.2GLFWAYY216 pKa = 10.73NGYY219 pKa = 9.42QLTDD223 pKa = 3.1QTIYY227 pKa = 10.83LSSDD231 pKa = 3.0SAALRR236 pKa = 11.84LEE238 pKa = 4.37ALGSPTFVYY247 pKa = 10.38KK248 pKa = 10.81VVGFGKK254 pKa = 10.16QKK256 pKa = 10.65VGLAVAIRR264 pKa = 11.84EE265 pKa = 4.06FQYY268 pKa = 11.38DD269 pKa = 4.07EE270 pKa = 4.07DD271 pKa = 4.19TGILTVSSLLITYY284 pKa = 10.28KK285 pKa = 10.72LDD287 pKa = 3.05IGKK290 pKa = 10.6GYY292 pKa = 10.7DD293 pKa = 3.01RR294 pKa = 11.84TKK296 pKa = 10.8FYY298 pKa = 11.34VDD300 pKa = 5.43DD301 pKa = 3.98IDD303 pKa = 5.37FGAGYY308 pKa = 8.36ITVLNGGIYY317 pKa = 10.44YY318 pKa = 10.64SGDD321 pKa = 3.24VPTFEE326 pKa = 5.26IPEE329 pKa = 4.3ACQPCPEE336 pKa = 4.12TAPWYY341 pKa = 8.92TDD343 pKa = 3.53SNPSDD348 pKa = 3.48VSTISDD354 pKa = 4.12DD355 pKa = 3.72NTSTVIDD362 pKa = 3.76QSSTEE367 pKa = 4.52PIPTTTDD374 pKa = 2.89NGSGSGSGSGSGSASATSTTSNDD397 pKa = 3.07GSGSDD402 pKa = 3.93GSGSGSGSASATSTTSNDD420 pKa = 3.07GSGSDD425 pKa = 3.93GSGSGSGSASATTTASDD442 pKa = 3.48NGSGSGSDD450 pKa = 3.34SGSGSGSDD458 pKa = 4.06SNNGSNSGSNSGSTNGSGSSNDD480 pKa = 3.21SSSNDD485 pKa = 3.03GSGSDD490 pKa = 3.93GSGSGSGSGSASATTTASDD509 pKa = 3.48NGSGSGSGSGSASATSTTSNDD530 pKa = 3.07GSGSDD535 pKa = 3.93GSGSGSGSASATTTASDD552 pKa = 3.48NGSGSGSDD560 pKa = 3.34SGSGSGSGSDD570 pKa = 4.06SNNGSNSGSNSGSTNGSGSSNDD592 pKa = 3.21SSSNDD597 pKa = 3.03GSGSDD602 pKa = 3.93GSGSGSGSGSASATTTASEE621 pKa = 4.23NGSGSGSGSGSASGSASGSASATGTDD647 pKa = 3.73NDD649 pKa = 4.1SSSGSGSDD657 pKa = 3.2SGSGSGSDD665 pKa = 4.06SNNGSNSSSNSGSTNGSGSNNDD687 pKa = 3.28SSSGSSSGSSSNNGSDD703 pKa = 4.25SSNTTYY709 pKa = 8.65PTIKK713 pKa = 7.94TTTKK717 pKa = 10.2AGASSTSATSSDD729 pKa = 3.0GSTDD733 pKa = 3.34AKK735 pKa = 8.35TTATASIYY743 pKa = 10.48TGGAALASAGTGLAAFAIGALLII766 pKa = 4.35
MM1 pKa = 6.79KK2 pKa = 9.98TSVVNVTILLACGVPLASALRR23 pKa = 11.84TVTSDD28 pKa = 3.22YY29 pKa = 11.59DD30 pKa = 4.15VISDD34 pKa = 3.77TVLDD38 pKa = 3.55VGGAIFNGNVQIEE51 pKa = 4.17EE52 pKa = 4.36DD53 pKa = 3.55KK54 pKa = 11.2FLALVSGTTEE64 pKa = 3.95YY65 pKa = 10.85FNKK68 pKa = 10.55DD69 pKa = 3.97LIVDD73 pKa = 4.17GALYY77 pKa = 10.13IGSKK81 pKa = 9.63IQTLGLDD88 pKa = 3.69VKK90 pKa = 11.28VLGSTQKK97 pKa = 10.02IQNTGTIVLNGHH109 pKa = 5.72NTTLPSTYY117 pKa = 9.46YY118 pKa = 9.79WVGEE122 pKa = 4.1SFKK125 pKa = 11.19NDD127 pKa = 3.02GEE129 pKa = 4.57IYY131 pKa = 10.14WIAQSSLIGSLYY143 pKa = 10.35EE144 pKa = 4.54IIPSSSFEE152 pKa = 4.4NNGLLSFTHH161 pKa = 5.9EE162 pKa = 4.22NGRR165 pKa = 11.84KK166 pKa = 9.93GGTLTLGNLGSAITNSGTICLKK188 pKa = 10.43GVVYY192 pKa = 9.93KK193 pKa = 10.31QSGMVSGEE201 pKa = 4.2SGCISIGDD209 pKa = 3.89EE210 pKa = 4.2GLFWAYY216 pKa = 10.73NGYY219 pKa = 9.42QLTDD223 pKa = 3.1QTIYY227 pKa = 10.83LSSDD231 pKa = 3.0SAALRR236 pKa = 11.84LEE238 pKa = 4.37ALGSPTFVYY247 pKa = 10.38KK248 pKa = 10.81VVGFGKK254 pKa = 10.16QKK256 pKa = 10.65VGLAVAIRR264 pKa = 11.84EE265 pKa = 4.06FQYY268 pKa = 11.38DD269 pKa = 4.07EE270 pKa = 4.07DD271 pKa = 4.19TGILTVSSLLITYY284 pKa = 10.28KK285 pKa = 10.72LDD287 pKa = 3.05IGKK290 pKa = 10.6GYY292 pKa = 10.7DD293 pKa = 3.01RR294 pKa = 11.84TKK296 pKa = 10.8FYY298 pKa = 11.34VDD300 pKa = 5.43DD301 pKa = 3.98IDD303 pKa = 5.37FGAGYY308 pKa = 8.36ITVLNGGIYY317 pKa = 10.44YY318 pKa = 10.64SGDD321 pKa = 3.24VPTFEE326 pKa = 5.26IPEE329 pKa = 4.3ACQPCPEE336 pKa = 4.12TAPWYY341 pKa = 8.92TDD343 pKa = 3.53SNPSDD348 pKa = 3.48VSTISDD354 pKa = 4.12DD355 pKa = 3.72NTSTVIDD362 pKa = 3.76QSSTEE367 pKa = 4.52PIPTTTDD374 pKa = 2.89NGSGSGSGSGSGSASATSTTSNDD397 pKa = 3.07GSGSDD402 pKa = 3.93GSGSGSGSASATSTTSNDD420 pKa = 3.07GSGSDD425 pKa = 3.93GSGSGSGSASATTTASDD442 pKa = 3.48NGSGSGSDD450 pKa = 3.34SGSGSGSDD458 pKa = 4.06SNNGSNSGSNSGSTNGSGSSNDD480 pKa = 3.21SSSNDD485 pKa = 3.03GSGSDD490 pKa = 3.93GSGSGSGSGSASATTTASDD509 pKa = 3.48NGSGSGSGSGSASATSTTSNDD530 pKa = 3.07GSGSDD535 pKa = 3.93GSGSGSGSASATTTASDD552 pKa = 3.48NGSGSGSDD560 pKa = 3.34SGSGSGSGSDD570 pKa = 4.06SNNGSNSGSNSGSTNGSGSSNDD592 pKa = 3.21SSSNDD597 pKa = 3.03GSGSDD602 pKa = 3.93GSGSGSGSGSASATTTASEE621 pKa = 4.23NGSGSGSGSGSASGSASGSASATGTDD647 pKa = 3.73NDD649 pKa = 4.1SSSGSGSDD657 pKa = 3.2SGSGSGSDD665 pKa = 4.06SNNGSNSSSNSGSTNGSGSNNDD687 pKa = 3.28SSSGSSSGSSSNNGSDD703 pKa = 4.25SSNTTYY709 pKa = 8.65PTIKK713 pKa = 7.94TTTKK717 pKa = 10.2AGASSTSATSSDD729 pKa = 3.0GSTDD733 pKa = 3.34AKK735 pKa = 8.35TTATASIYY743 pKa = 10.48TGGAALASAGTGLAAFAIGALLII766 pKa = 4.35
Molecular weight: 73.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6CPF8|Q6CPF8_KLULA Elongator complex protein 1 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=KLLA0_E05237g PE=3 SV=1
MM1 pKa = 7.83AKK3 pKa = 8.75RR4 pKa = 11.84TKK6 pKa = 10.44KK7 pKa = 10.64VGITGKK13 pKa = 10.38YY14 pKa = 7.24GVRR17 pKa = 11.84YY18 pKa = 9.34GSSLRR23 pKa = 11.84RR24 pKa = 11.84QVKK27 pKa = 9.69KK28 pKa = 11.2LEE30 pKa = 4.24VQQHH34 pKa = 4.32ATYY37 pKa = 10.74NCSFCGKK44 pKa = 7.69TCVKK48 pKa = 10.36RR49 pKa = 11.84GAAGVWSCSSCNRR62 pKa = 11.84TIAGGAYY69 pKa = 8.84TLSTAAAATVRR80 pKa = 11.84STIRR84 pKa = 11.84RR85 pKa = 11.84LRR87 pKa = 11.84DD88 pKa = 3.06MAEE91 pKa = 3.6AA92 pKa = 4.27
MM1 pKa = 7.83AKK3 pKa = 8.75RR4 pKa = 11.84TKK6 pKa = 10.44KK7 pKa = 10.64VGITGKK13 pKa = 10.38YY14 pKa = 7.24GVRR17 pKa = 11.84YY18 pKa = 9.34GSSLRR23 pKa = 11.84RR24 pKa = 11.84QVKK27 pKa = 9.69KK28 pKa = 11.2LEE30 pKa = 4.24VQQHH34 pKa = 4.32ATYY37 pKa = 10.74NCSFCGKK44 pKa = 7.69TCVKK48 pKa = 10.36RR49 pKa = 11.84GAAGVWSCSSCNRR62 pKa = 11.84TIAGGAYY69 pKa = 8.84TLSTAAAATVRR80 pKa = 11.84STIRR84 pKa = 11.84RR85 pKa = 11.84LRR87 pKa = 11.84DD88 pKa = 3.06MAEE91 pKa = 3.6AA92 pKa = 4.27
Molecular weight: 9.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2461417 |
39 |
4915 |
485.4 |
54.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.595 ± 0.03 | 1.236 ± 0.012 |
6.057 ± 0.026 | 6.699 ± 0.038 |
4.384 ± 0.025 | 5.097 ± 0.03 |
2.188 ± 0.014 | 6.355 ± 0.027 |
7.079 ± 0.031 | 9.702 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.08 ± 0.013 | 5.565 ± 0.023 |
4.303 ± 0.027 | 4.2 ± 0.03 |
4.352 ± 0.022 | 8.83 ± 0.041 |
5.83 ± 0.031 | 6.001 ± 0.023 |
1.089 ± 0.011 | 3.358 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
