
Wolbachia endosymbiont wVitA of Nasonia vitripennis phage WOVitA1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
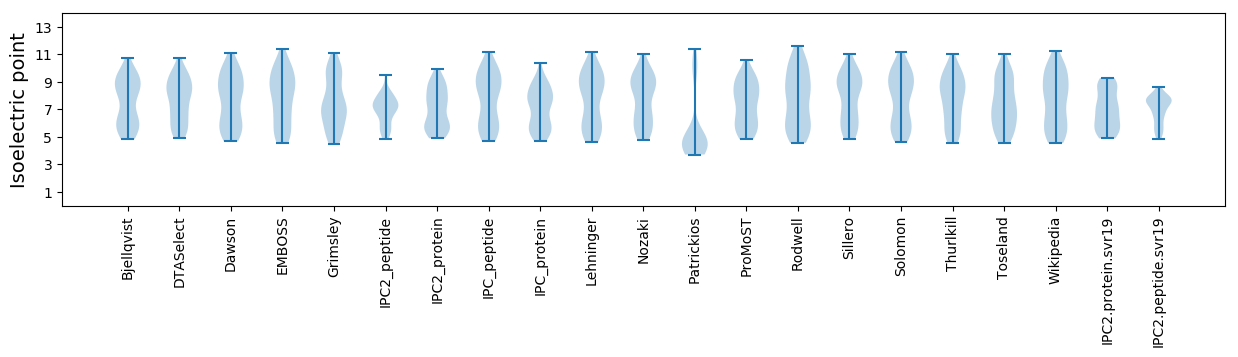
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
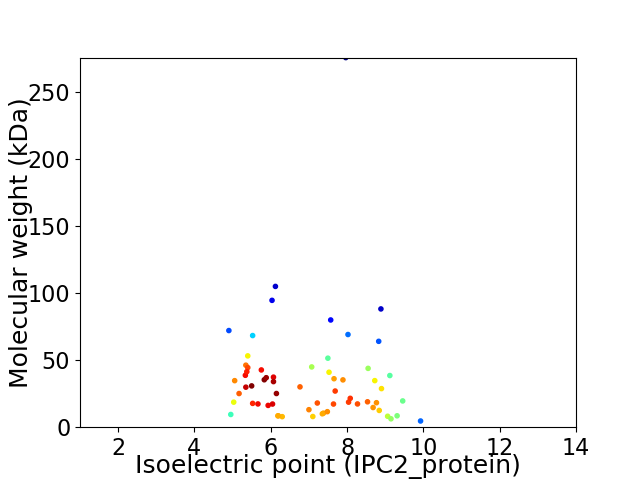
Virtual 2D-PAGE plot for 63 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E9P5Y1|E9P5Y1_9CAUD Putative virulence protein VrlC.2 OS=Wolbachia endosymbiont wVitA of Nasonia vitripennis phage WOVitA1 OX=949124 PE=4 SV=1
MM1 pKa = 7.4TLNSYY6 pKa = 9.26YY7 pKa = 11.01NRR9 pKa = 11.84FNPDD13 pKa = 2.6KK14 pKa = 9.93EE15 pKa = 4.45YY16 pKa = 11.14EE17 pKa = 4.1KK18 pKa = 11.26SLFLAGRR25 pKa = 11.84GLQSAEE31 pKa = 3.83LNEE34 pKa = 4.2TQEE37 pKa = 4.28YY38 pKa = 10.71ALSKK42 pKa = 10.85LKK44 pKa = 11.13GIGDD48 pKa = 4.62AIFRR52 pKa = 11.84DD53 pKa = 3.75GDD55 pKa = 3.88VITGSNCIIDD65 pKa = 4.07RR66 pKa = 11.84EE67 pKa = 4.45TGKK70 pKa = 8.52VTLEE74 pKa = 3.66SGKK77 pKa = 10.1IYY79 pKa = 10.61LRR81 pKa = 11.84GAVRR85 pKa = 11.84KK86 pKa = 9.17VEE88 pKa = 4.08KK89 pKa = 10.55EE90 pKa = 3.86EE91 pKa = 3.91FVIPLSTTVRR101 pKa = 11.84IGVYY105 pKa = 9.22YY106 pKa = 10.39VEE108 pKa = 4.3STITEE113 pKa = 4.26LEE115 pKa = 4.21DD116 pKa = 3.35EE117 pKa = 4.55NLRR120 pKa = 11.84DD121 pKa = 3.55PAIGTRR127 pKa = 11.84NYY129 pKa = 10.54QEE131 pKa = 3.78VGAARR136 pKa = 11.84LKK138 pKa = 10.67ISTIWGYY145 pKa = 7.57QAEE148 pKa = 4.4GLSPRR153 pKa = 11.84FSEE156 pKa = 4.7GEE158 pKa = 3.83FYY160 pKa = 10.49PIYY163 pKa = 10.54NIEE166 pKa = 4.08NGVLIEE172 pKa = 4.85HH173 pKa = 6.79SPPPQANIVTTALARR188 pKa = 11.84YY189 pKa = 9.06DD190 pKa = 3.57KK191 pKa = 9.9EE192 pKa = 4.82ANGSYY197 pKa = 9.87VVNGLEE203 pKa = 4.41VMFLQKK209 pKa = 11.01EE210 pKa = 4.04EE211 pKa = 4.63GEE213 pKa = 4.33GGKK216 pKa = 10.4KK217 pKa = 9.7IFVINEE223 pKa = 3.86GKK225 pKa = 10.3AHH227 pKa = 5.83VDD229 pKa = 3.42GYY231 pKa = 10.76EE232 pKa = 3.51IEE234 pKa = 4.62LPHH237 pKa = 7.14SIRR240 pKa = 11.84ISFDD244 pKa = 3.15EE245 pKa = 4.85DD246 pKa = 3.65PDD248 pKa = 3.74IKK250 pKa = 10.84SVEE253 pKa = 4.35SEE255 pKa = 3.84PHH257 pKa = 5.03TFQPNSQRR265 pKa = 11.84VMEE268 pKa = 4.64LKK270 pKa = 11.15VNDD273 pKa = 4.17FPISEE278 pKa = 4.18IKK280 pKa = 10.58KK281 pKa = 10.0VDD283 pKa = 2.99ITIQKK288 pKa = 8.83TITITHH294 pKa = 6.78GSYY297 pKa = 10.42SGAIDD302 pKa = 5.63PIPDD306 pKa = 3.53SAVLEE311 pKa = 4.54IIQAKK316 pKa = 8.26QGNVIYY322 pKa = 10.13EE323 pKa = 3.81NSVDD327 pKa = 3.77YY328 pKa = 10.77KK329 pKa = 11.34LNAGNVDD336 pKa = 2.72WSLPGKK342 pKa = 9.9EE343 pKa = 4.11PAPGSSYY350 pKa = 10.72QITYY354 pKa = 9.83RR355 pKa = 11.84CRR357 pKa = 11.84THH359 pKa = 6.91VSPEE363 pKa = 4.38DD364 pKa = 3.55ISEE367 pKa = 4.21EE368 pKa = 4.29GCKK371 pKa = 10.67VKK373 pKa = 10.77GAVDD377 pKa = 3.53NSLVLIDD384 pKa = 4.7YY385 pKa = 6.81TWKK388 pKa = 9.73MPRR391 pKa = 11.84FDD393 pKa = 6.42LITIDD398 pKa = 3.12SKK400 pKa = 11.48GVVRR404 pKa = 11.84RR405 pKa = 11.84IKK407 pKa = 10.75GIAHH411 pKa = 7.37PWRR414 pKa = 11.84PSMPRR419 pKa = 11.84APSGQLLLCYY429 pKa = 10.21IHH431 pKa = 5.87QTWKK435 pKa = 10.49NGEE438 pKa = 3.99GVKK441 pKa = 9.94IVNNAIHH448 pKa = 6.62AVPMNEE454 pKa = 3.93LEE456 pKa = 4.22AMKK459 pKa = 10.52KK460 pKa = 10.43GINDD464 pKa = 4.22LYY466 pKa = 11.47ALVAEE471 pKa = 4.41EE472 pKa = 4.7RR473 pKa = 11.84LRR475 pKa = 11.84SDD477 pKa = 3.24ANSRR481 pKa = 11.84EE482 pKa = 4.13PTTKK486 pKa = 10.14KK487 pKa = 10.7GVFVDD492 pKa = 4.65SFFDD496 pKa = 4.97DD497 pKa = 3.92DD498 pKa = 4.83MRR500 pKa = 11.84DD501 pKa = 3.34QGISQSAAIVNKK513 pKa = 10.03EE514 pKa = 4.43LILPIDD520 pKa = 3.68VTIADD525 pKa = 3.53IDD527 pKa = 4.11GGEE530 pKa = 4.06KK531 pKa = 9.92TYY533 pKa = 11.02LLPYY537 pKa = 9.52EE538 pKa = 4.66LEE540 pKa = 4.31PVLEE544 pKa = 4.23QLLQTKK550 pKa = 9.14GEE552 pKa = 4.45KK553 pKa = 9.8INPYY557 pKa = 9.47QAFDD561 pKa = 3.98PVPAQITLNKK571 pKa = 10.41NIDD574 pKa = 3.08HH575 pKa = 5.88WTEE578 pKa = 3.68VTTNWKK584 pKa = 10.57SPVTRR589 pKa = 11.84VFNVKK594 pKa = 8.07EE595 pKa = 4.29TTEE598 pKa = 4.3LLSSTSYY605 pKa = 9.44EE606 pKa = 3.91AEE608 pKa = 4.02FMRR611 pKa = 11.84EE612 pKa = 3.74AVQDD616 pKa = 3.71FEE618 pKa = 5.51IEE620 pKa = 4.04GFEE623 pKa = 4.03PNEE626 pKa = 3.67RR627 pKa = 11.84LKK629 pKa = 10.66EE630 pKa = 3.96IKK632 pKa = 9.95FDD634 pKa = 3.63GMSIQPTAA642 pKa = 3.52
MM1 pKa = 7.4TLNSYY6 pKa = 9.26YY7 pKa = 11.01NRR9 pKa = 11.84FNPDD13 pKa = 2.6KK14 pKa = 9.93EE15 pKa = 4.45YY16 pKa = 11.14EE17 pKa = 4.1KK18 pKa = 11.26SLFLAGRR25 pKa = 11.84GLQSAEE31 pKa = 3.83LNEE34 pKa = 4.2TQEE37 pKa = 4.28YY38 pKa = 10.71ALSKK42 pKa = 10.85LKK44 pKa = 11.13GIGDD48 pKa = 4.62AIFRR52 pKa = 11.84DD53 pKa = 3.75GDD55 pKa = 3.88VITGSNCIIDD65 pKa = 4.07RR66 pKa = 11.84EE67 pKa = 4.45TGKK70 pKa = 8.52VTLEE74 pKa = 3.66SGKK77 pKa = 10.1IYY79 pKa = 10.61LRR81 pKa = 11.84GAVRR85 pKa = 11.84KK86 pKa = 9.17VEE88 pKa = 4.08KK89 pKa = 10.55EE90 pKa = 3.86EE91 pKa = 3.91FVIPLSTTVRR101 pKa = 11.84IGVYY105 pKa = 9.22YY106 pKa = 10.39VEE108 pKa = 4.3STITEE113 pKa = 4.26LEE115 pKa = 4.21DD116 pKa = 3.35EE117 pKa = 4.55NLRR120 pKa = 11.84DD121 pKa = 3.55PAIGTRR127 pKa = 11.84NYY129 pKa = 10.54QEE131 pKa = 3.78VGAARR136 pKa = 11.84LKK138 pKa = 10.67ISTIWGYY145 pKa = 7.57QAEE148 pKa = 4.4GLSPRR153 pKa = 11.84FSEE156 pKa = 4.7GEE158 pKa = 3.83FYY160 pKa = 10.49PIYY163 pKa = 10.54NIEE166 pKa = 4.08NGVLIEE172 pKa = 4.85HH173 pKa = 6.79SPPPQANIVTTALARR188 pKa = 11.84YY189 pKa = 9.06DD190 pKa = 3.57KK191 pKa = 9.9EE192 pKa = 4.82ANGSYY197 pKa = 9.87VVNGLEE203 pKa = 4.41VMFLQKK209 pKa = 11.01EE210 pKa = 4.04EE211 pKa = 4.63GEE213 pKa = 4.33GGKK216 pKa = 10.4KK217 pKa = 9.7IFVINEE223 pKa = 3.86GKK225 pKa = 10.3AHH227 pKa = 5.83VDD229 pKa = 3.42GYY231 pKa = 10.76EE232 pKa = 3.51IEE234 pKa = 4.62LPHH237 pKa = 7.14SIRR240 pKa = 11.84ISFDD244 pKa = 3.15EE245 pKa = 4.85DD246 pKa = 3.65PDD248 pKa = 3.74IKK250 pKa = 10.84SVEE253 pKa = 4.35SEE255 pKa = 3.84PHH257 pKa = 5.03TFQPNSQRR265 pKa = 11.84VMEE268 pKa = 4.64LKK270 pKa = 11.15VNDD273 pKa = 4.17FPISEE278 pKa = 4.18IKK280 pKa = 10.58KK281 pKa = 10.0VDD283 pKa = 2.99ITIQKK288 pKa = 8.83TITITHH294 pKa = 6.78GSYY297 pKa = 10.42SGAIDD302 pKa = 5.63PIPDD306 pKa = 3.53SAVLEE311 pKa = 4.54IIQAKK316 pKa = 8.26QGNVIYY322 pKa = 10.13EE323 pKa = 3.81NSVDD327 pKa = 3.77YY328 pKa = 10.77KK329 pKa = 11.34LNAGNVDD336 pKa = 2.72WSLPGKK342 pKa = 9.9EE343 pKa = 4.11PAPGSSYY350 pKa = 10.72QITYY354 pKa = 9.83RR355 pKa = 11.84CRR357 pKa = 11.84THH359 pKa = 6.91VSPEE363 pKa = 4.38DD364 pKa = 3.55ISEE367 pKa = 4.21EE368 pKa = 4.29GCKK371 pKa = 10.67VKK373 pKa = 10.77GAVDD377 pKa = 3.53NSLVLIDD384 pKa = 4.7YY385 pKa = 6.81TWKK388 pKa = 9.73MPRR391 pKa = 11.84FDD393 pKa = 6.42LITIDD398 pKa = 3.12SKK400 pKa = 11.48GVVRR404 pKa = 11.84RR405 pKa = 11.84IKK407 pKa = 10.75GIAHH411 pKa = 7.37PWRR414 pKa = 11.84PSMPRR419 pKa = 11.84APSGQLLLCYY429 pKa = 10.21IHH431 pKa = 5.87QTWKK435 pKa = 10.49NGEE438 pKa = 3.99GVKK441 pKa = 9.94IVNNAIHH448 pKa = 6.62AVPMNEE454 pKa = 3.93LEE456 pKa = 4.22AMKK459 pKa = 10.52KK460 pKa = 10.43GINDD464 pKa = 4.22LYY466 pKa = 11.47ALVAEE471 pKa = 4.41EE472 pKa = 4.7RR473 pKa = 11.84LRR475 pKa = 11.84SDD477 pKa = 3.24ANSRR481 pKa = 11.84EE482 pKa = 4.13PTTKK486 pKa = 10.14KK487 pKa = 10.7GVFVDD492 pKa = 4.65SFFDD496 pKa = 4.97DD497 pKa = 3.92DD498 pKa = 4.83MRR500 pKa = 11.84DD501 pKa = 3.34QGISQSAAIVNKK513 pKa = 10.03EE514 pKa = 4.43LILPIDD520 pKa = 3.68VTIADD525 pKa = 3.53IDD527 pKa = 4.11GGEE530 pKa = 4.06KK531 pKa = 9.92TYY533 pKa = 11.02LLPYY537 pKa = 9.52EE538 pKa = 4.66LEE540 pKa = 4.31PVLEE544 pKa = 4.23QLLQTKK550 pKa = 9.14GEE552 pKa = 4.45KK553 pKa = 9.8INPYY557 pKa = 9.47QAFDD561 pKa = 3.98PVPAQITLNKK571 pKa = 10.41NIDD574 pKa = 3.08HH575 pKa = 5.88WTEE578 pKa = 3.68VTTNWKK584 pKa = 10.57SPVTRR589 pKa = 11.84VFNVKK594 pKa = 8.07EE595 pKa = 4.29TTEE598 pKa = 4.3LLSSTSYY605 pKa = 9.44EE606 pKa = 3.91AEE608 pKa = 4.02FMRR611 pKa = 11.84EE612 pKa = 3.74AVQDD616 pKa = 3.71FEE618 pKa = 5.51IEE620 pKa = 4.04GFEE623 pKa = 4.03PNEE626 pKa = 3.67RR627 pKa = 11.84LKK629 pKa = 10.66EE630 pKa = 3.96IKK632 pKa = 9.95FDD634 pKa = 3.63GMSIQPTAA642 pKa = 3.52
Molecular weight: 72.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E9P5X2|E9P5X2_9CAUD Uncharacterized protein OS=Wolbachia endosymbiont wVitA of Nasonia vitripennis phage WOVitA1 OX=949124 PE=4 SV=1
MM1 pKa = 7.54LRR3 pKa = 11.84PFGKK7 pKa = 10.08IKK9 pKa = 10.37IFAKK13 pKa = 10.36NSIVFKK19 pKa = 10.97LLMFSCFIYY28 pKa = 9.82TLRR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.57GFLKK37 pKa = 10.64KK38 pKa = 10.65GG39 pKa = 3.39
MM1 pKa = 7.54LRR3 pKa = 11.84PFGKK7 pKa = 10.08IKK9 pKa = 10.37IFAKK13 pKa = 10.36NSIVFKK19 pKa = 10.97LLMFSCFIYY28 pKa = 9.82TLRR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.57GFLKK37 pKa = 10.64KK38 pKa = 10.65GG39 pKa = 3.39
Molecular weight: 4.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
20107 |
39 |
2474 |
319.2 |
35.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.709 ± 0.332 | 1.025 ± 0.125 |
5.003 ± 0.156 | 8.087 ± 0.373 |
3.481 ± 0.175 | 6.102 ± 0.22 |
2.079 ± 0.203 | 8.092 ± 0.271 |
8.863 ± 0.288 | 9.723 ± 0.37 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.726 ± 0.143 | 6.058 ± 0.446 |
2.989 ± 0.212 | 3.098 ± 0.134 |
4.6 ± 0.3 | 6.729 ± 0.221 |
4.958 ± 0.192 | 6.495 ± 0.181 |
0.935 ± 0.142 | 3.248 ± 0.166 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
