
Natronorubrum tibetense GA33
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natronorubrum; Natronorubrum tibetense
Average proteome isoelectric point is 4.8
Get precalculated fractions of proteins
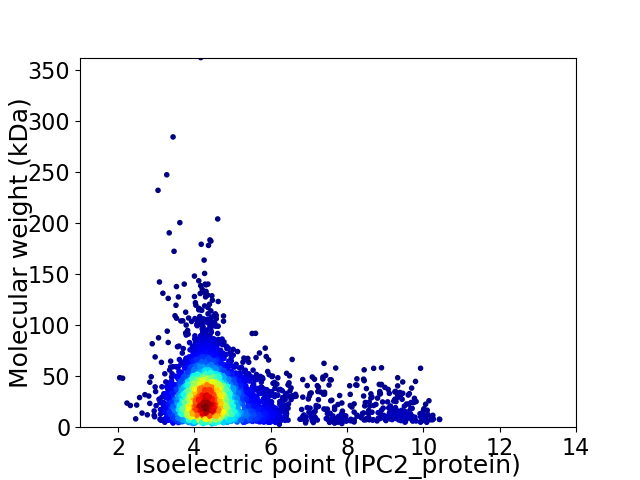
Virtual 2D-PAGE plot for 4671 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L9VRY1|L9VRY1_9EURY Basic membrane lipoprotein OS=Natronorubrum tibetense GA33 OX=1114856 GN=C496_13756 PE=4 SV=1
MM1 pKa = 7.31GASDD5 pKa = 4.24TIAAYY10 pKa = 10.27FGFDD14 pKa = 3.18EE15 pKa = 5.59HH16 pKa = 6.12EE17 pKa = 3.78TDD19 pKa = 4.43YY20 pKa = 10.42RR21 pKa = 11.84TEE23 pKa = 3.94TLAGVTTFLAMAYY36 pKa = 9.32IIVVNPAILAEE47 pKa = 4.97AITIDD52 pKa = 4.42GYY54 pKa = 9.96TDD56 pKa = 3.0AQTRR60 pKa = 11.84DD61 pKa = 3.56VIAIATILASVLAIGVMAFWANRR84 pKa = 11.84PFGLAPGMGLNAFFAFTVVIILGVPWEE111 pKa = 4.16LALAAVFVEE120 pKa = 4.81GIIFIALTAVGARR133 pKa = 11.84KK134 pKa = 9.95YY135 pKa = 10.35IIEE138 pKa = 4.79LFPEE142 pKa = 4.31PVKK145 pKa = 10.53FAVGAGIGVFLLFLGLQEE163 pKa = 4.19MEE165 pKa = 4.36VVVGYY170 pKa = 10.12DD171 pKa = 3.22ATLLTLGNVLEE182 pKa = 4.61SPVAALSLTGLALTFFLYY200 pKa = 10.82ARR202 pKa = 11.84GIRR205 pKa = 11.84GSIIIGILSTAVAGWLLTLLDD226 pKa = 4.02VVSPGSLVPEE236 pKa = 4.34PAYY239 pKa = 10.61DD240 pKa = 3.27QATNDD245 pKa = 3.93GLFSMLLSVEE255 pKa = 3.92YY256 pKa = 10.4DD257 pKa = 3.15ITPLISGFIDD267 pKa = 3.91GLGMMADD274 pKa = 4.11DD275 pKa = 4.55PLVFALVVFTFFFVDD290 pKa = 4.8FFDD293 pKa = 3.96TAGTLIGVSQIGGFLDD309 pKa = 4.87DD310 pKa = 5.36EE311 pKa = 5.43GDD313 pKa = 3.51LPEE316 pKa = 4.97IEE318 pKa = 4.57RR319 pKa = 11.84PLMADD324 pKa = 2.96AVGTTFGAMIGTSTVTTYY342 pKa = 10.35IEE344 pKa = 4.22SSTGIEE350 pKa = 3.78EE351 pKa = 4.36GGRR354 pKa = 11.84TGFTALIVGVLFLAALLFVPLMRR377 pKa = 11.84AVPQYY382 pKa = 9.06ATYY385 pKa = 10.05IALVVVGIIMLQGVADD401 pKa = 4.96IDD403 pKa = 3.65WQDD406 pKa = 3.6PVWAISAGLTITIMPLTASIANGLAAGIISYY437 pKa = 9.36PLIKK441 pKa = 10.31AAIGEE446 pKa = 4.25ARR448 pKa = 11.84DD449 pKa = 3.57VSPGQWVLAVLFVGYY464 pKa = 8.81FAIYY468 pKa = 9.22FAVEE472 pKa = 3.93AGQITFF478 pKa = 3.94
MM1 pKa = 7.31GASDD5 pKa = 4.24TIAAYY10 pKa = 10.27FGFDD14 pKa = 3.18EE15 pKa = 5.59HH16 pKa = 6.12EE17 pKa = 3.78TDD19 pKa = 4.43YY20 pKa = 10.42RR21 pKa = 11.84TEE23 pKa = 3.94TLAGVTTFLAMAYY36 pKa = 9.32IIVVNPAILAEE47 pKa = 4.97AITIDD52 pKa = 4.42GYY54 pKa = 9.96TDD56 pKa = 3.0AQTRR60 pKa = 11.84DD61 pKa = 3.56VIAIATILASVLAIGVMAFWANRR84 pKa = 11.84PFGLAPGMGLNAFFAFTVVIILGVPWEE111 pKa = 4.16LALAAVFVEE120 pKa = 4.81GIIFIALTAVGARR133 pKa = 11.84KK134 pKa = 9.95YY135 pKa = 10.35IIEE138 pKa = 4.79LFPEE142 pKa = 4.31PVKK145 pKa = 10.53FAVGAGIGVFLLFLGLQEE163 pKa = 4.19MEE165 pKa = 4.36VVVGYY170 pKa = 10.12DD171 pKa = 3.22ATLLTLGNVLEE182 pKa = 4.61SPVAALSLTGLALTFFLYY200 pKa = 10.82ARR202 pKa = 11.84GIRR205 pKa = 11.84GSIIIGILSTAVAGWLLTLLDD226 pKa = 4.02VVSPGSLVPEE236 pKa = 4.34PAYY239 pKa = 10.61DD240 pKa = 3.27QATNDD245 pKa = 3.93GLFSMLLSVEE255 pKa = 3.92YY256 pKa = 10.4DD257 pKa = 3.15ITPLISGFIDD267 pKa = 3.91GLGMMADD274 pKa = 4.11DD275 pKa = 4.55PLVFALVVFTFFFVDD290 pKa = 4.8FFDD293 pKa = 3.96TAGTLIGVSQIGGFLDD309 pKa = 4.87DD310 pKa = 5.36EE311 pKa = 5.43GDD313 pKa = 3.51LPEE316 pKa = 4.97IEE318 pKa = 4.57RR319 pKa = 11.84PLMADD324 pKa = 2.96AVGTTFGAMIGTSTVTTYY342 pKa = 10.35IEE344 pKa = 4.22SSTGIEE350 pKa = 3.78EE351 pKa = 4.36GGRR354 pKa = 11.84TGFTALIVGVLFLAALLFVPLMRR377 pKa = 11.84AVPQYY382 pKa = 9.06ATYY385 pKa = 10.05IALVVVGIIMLQGVADD401 pKa = 4.96IDD403 pKa = 3.65WQDD406 pKa = 3.6PVWAISAGLTITIMPLTASIANGLAAGIISYY437 pKa = 9.36PLIKK441 pKa = 10.31AAIGEE446 pKa = 4.25ARR448 pKa = 11.84DD449 pKa = 3.57VSPGQWVLAVLFVGYY464 pKa = 8.81FAIYY468 pKa = 9.22FAVEE472 pKa = 3.93AGQITFF478 pKa = 3.94
Molecular weight: 50.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L9VPD0|L9VPD0_9EURY Uncharacterized protein OS=Natronorubrum tibetense GA33 OX=1114856 GN=C496_16057 PE=3 SV=1
MM1 pKa = 7.55SKK3 pKa = 10.76GEE5 pKa = 3.81KK6 pKa = 10.11AIWIWAPIIAKK17 pKa = 9.69KK18 pKa = 10.4CPKK21 pKa = 9.74CGNSSSYY28 pKa = 10.68HH29 pKa = 5.33EE30 pKa = 5.18RR31 pKa = 11.84SDD33 pKa = 3.87CEE35 pKa = 4.01YY36 pKa = 11.3DD37 pKa = 3.32EE38 pKa = 4.92TEE40 pKa = 3.8PDD42 pKa = 3.25EE43 pKa = 4.38WNKK46 pKa = 10.67GLVGFRR52 pKa = 11.84PAPVFDD58 pKa = 3.97ISQTKK63 pKa = 10.39GEE65 pKa = 4.55SLPEE69 pKa = 4.13LEE71 pKa = 5.08TVLSLPPLRR80 pKa = 11.84EE81 pKa = 3.51LEE83 pKa = 4.09QRR85 pKa = 11.84VDD87 pKa = 3.27RR88 pKa = 11.84HH89 pKa = 5.55SLGVGSSAGRR99 pKa = 11.84RR100 pKa = 11.84LCGCARR106 pKa = 11.84GGHH109 pKa = 5.66LRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84PRR115 pKa = 11.84LRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84DD120 pKa = 3.76PRR122 pKa = 11.84GGAPPSRR129 pKa = 11.84VRR131 pKa = 11.84VPRR134 pKa = 11.84VRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84GLCSRR143 pKa = 4.63
MM1 pKa = 7.55SKK3 pKa = 10.76GEE5 pKa = 3.81KK6 pKa = 10.11AIWIWAPIIAKK17 pKa = 9.69KK18 pKa = 10.4CPKK21 pKa = 9.74CGNSSSYY28 pKa = 10.68HH29 pKa = 5.33EE30 pKa = 5.18RR31 pKa = 11.84SDD33 pKa = 3.87CEE35 pKa = 4.01YY36 pKa = 11.3DD37 pKa = 3.32EE38 pKa = 4.92TEE40 pKa = 3.8PDD42 pKa = 3.25EE43 pKa = 4.38WNKK46 pKa = 10.67GLVGFRR52 pKa = 11.84PAPVFDD58 pKa = 3.97ISQTKK63 pKa = 10.39GEE65 pKa = 4.55SLPEE69 pKa = 4.13LEE71 pKa = 5.08TVLSLPPLRR80 pKa = 11.84EE81 pKa = 3.51LEE83 pKa = 4.09QRR85 pKa = 11.84VDD87 pKa = 3.27RR88 pKa = 11.84HH89 pKa = 5.55SLGVGSSAGRR99 pKa = 11.84RR100 pKa = 11.84LCGCARR106 pKa = 11.84GGHH109 pKa = 5.66LRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84PRR115 pKa = 11.84LRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84DD120 pKa = 3.76PRR122 pKa = 11.84GGAPPSRR129 pKa = 11.84VRR131 pKa = 11.84VPRR134 pKa = 11.84VRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84GLCSRR143 pKa = 4.63
Molecular weight: 16.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1348622 |
27 |
3252 |
288.7 |
31.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.171 ± 0.051 | 0.773 ± 0.011 |
8.714 ± 0.058 | 9.268 ± 0.055 |
3.295 ± 0.025 | 8.198 ± 0.036 |
2.048 ± 0.019 | 4.597 ± 0.032 |
1.797 ± 0.022 | 8.867 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.743 ± 0.016 | 2.407 ± 0.021 |
4.531 ± 0.024 | 2.548 ± 0.023 |
6.301 ± 0.044 | 5.765 ± 0.029 |
6.547 ± 0.033 | 8.547 ± 0.038 |
1.138 ± 0.014 | 2.745 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
