
Hubei tombus-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.23
Get precalculated fractions of proteins
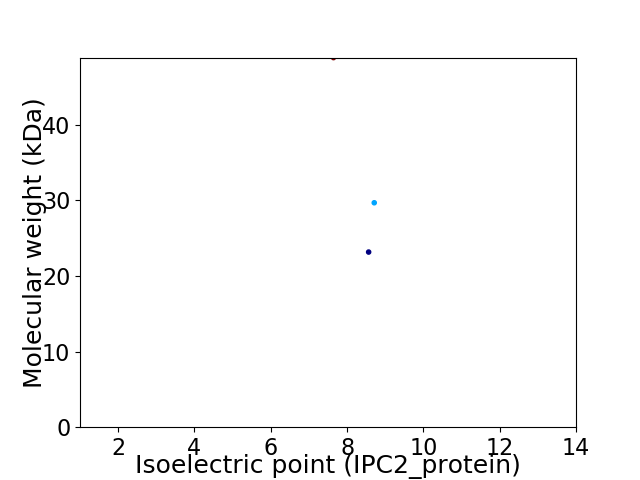
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KGN2|A0A1L3KGN2_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 6 OX=1923293 PE=4 SV=1
MM1 pKa = 7.38TSVKK5 pKa = 10.64DD6 pKa = 3.55SLRR9 pKa = 11.84KK10 pKa = 9.29FLPSTAPISHH20 pKa = 7.34DD21 pKa = 3.39AFVSGYY27 pKa = 10.02KK28 pKa = 9.94GRR30 pKa = 11.84KK31 pKa = 7.76KK32 pKa = 10.39EE33 pKa = 3.95LYY35 pKa = 9.81QRR37 pKa = 11.84ALDD40 pKa = 4.31EE41 pKa = 4.32IRR43 pKa = 11.84ATEE46 pKa = 4.13SNLEE50 pKa = 3.77NDD52 pKa = 2.77SRR54 pKa = 11.84LNVFVKK60 pKa = 10.46FEE62 pKa = 4.31KK63 pKa = 9.94TDD65 pKa = 3.25HH66 pKa = 5.27TTKK69 pKa = 10.43KK70 pKa = 10.61DD71 pKa = 3.31PVPRR75 pKa = 11.84VISPRR80 pKa = 11.84DD81 pKa = 3.17PKK83 pKa = 11.03FNIRR87 pKa = 11.84VGRR90 pKa = 11.84YY91 pKa = 7.85LKK93 pKa = 10.35PLEE96 pKa = 3.97EE97 pKa = 4.56RR98 pKa = 11.84LFKK101 pKa = 11.1SLGKK105 pKa = 10.59LFGHH109 pKa = 5.69KK110 pKa = 8.79TVLKK114 pKa = 10.0GVNSQRR120 pKa = 11.84SAEE123 pKa = 3.97LLKK126 pKa = 10.69EE127 pKa = 3.6KK128 pKa = 10.41WEE130 pKa = 4.17MYY132 pKa = 9.84RR133 pKa = 11.84NPVAIGLDD141 pKa = 3.21ASRR144 pKa = 11.84FDD146 pKa = 3.34QHH148 pKa = 8.13VSIDD152 pKa = 3.5ALRR155 pKa = 11.84WEE157 pKa = 4.0HH158 pKa = 7.12SIYY161 pKa = 10.83LEE163 pKa = 4.16CFPQRR168 pKa = 11.84KK169 pKa = 7.59HH170 pKa = 5.33QEE172 pKa = 3.57RR173 pKa = 11.84LRR175 pKa = 11.84RR176 pKa = 11.84LLKK179 pKa = 10.44LQEE182 pKa = 4.56LNSCTGYY189 pKa = 9.13TPDD192 pKa = 5.1GKK194 pKa = 10.79LKK196 pKa = 10.81YY197 pKa = 8.99NVKK200 pKa = 7.88GTRR203 pKa = 11.84MSGDD207 pKa = 3.37MNTSLGNCVLMCSMIKK223 pKa = 10.19AYY225 pKa = 10.77LMSKK229 pKa = 10.6GIDD232 pKa = 3.24GQLANNGDD240 pKa = 3.88DD241 pKa = 3.47CVVFMEE247 pKa = 5.83KK248 pKa = 10.31SDD250 pKa = 4.23LLNFSDD256 pKa = 5.87GLYY259 pKa = 10.85DD260 pKa = 3.33WFLALGFNMAIEE272 pKa = 4.19EE273 pKa = 4.26PVFEE277 pKa = 4.23FEE279 pKa = 5.54QIEE282 pKa = 4.56FCQTKK287 pKa = 9.6PVFDD291 pKa = 3.71GEE293 pKa = 4.2YY294 pKa = 9.9WIMCRR299 pKa = 11.84NPHH302 pKa = 4.89TAINKK307 pKa = 9.47DD308 pKa = 3.4SVMLKK313 pKa = 10.24PFSCSSEE320 pKa = 3.6FRR322 pKa = 11.84GWLDD326 pKa = 3.33AVGTGGLAMAGGIPIFQEE344 pKa = 4.83LYY346 pKa = 9.16RR347 pKa = 11.84TYY349 pKa = 10.53QKK351 pKa = 10.62CGQQRR356 pKa = 11.84KK357 pKa = 9.04ISEE360 pKa = 4.62DD361 pKa = 3.43LLPWSFTNLSKK372 pKa = 11.06GMTRR376 pKa = 11.84QYY378 pKa = 10.57GTVSPAARR386 pKa = 11.84ASFYY390 pKa = 10.01WAFDD394 pKa = 3.26ITPDD398 pKa = 3.56EE399 pKa = 4.4QIALEE404 pKa = 4.78DD405 pKa = 4.47YY406 pKa = 10.67YY407 pKa = 11.63SSTKK411 pKa = 9.97IVSSLGPYY419 pKa = 8.85EE420 pKa = 3.91PRR422 pKa = 11.84GVFAEE427 pKa = 4.2
MM1 pKa = 7.38TSVKK5 pKa = 10.64DD6 pKa = 3.55SLRR9 pKa = 11.84KK10 pKa = 9.29FLPSTAPISHH20 pKa = 7.34DD21 pKa = 3.39AFVSGYY27 pKa = 10.02KK28 pKa = 9.94GRR30 pKa = 11.84KK31 pKa = 7.76KK32 pKa = 10.39EE33 pKa = 3.95LYY35 pKa = 9.81QRR37 pKa = 11.84ALDD40 pKa = 4.31EE41 pKa = 4.32IRR43 pKa = 11.84ATEE46 pKa = 4.13SNLEE50 pKa = 3.77NDD52 pKa = 2.77SRR54 pKa = 11.84LNVFVKK60 pKa = 10.46FEE62 pKa = 4.31KK63 pKa = 9.94TDD65 pKa = 3.25HH66 pKa = 5.27TTKK69 pKa = 10.43KK70 pKa = 10.61DD71 pKa = 3.31PVPRR75 pKa = 11.84VISPRR80 pKa = 11.84DD81 pKa = 3.17PKK83 pKa = 11.03FNIRR87 pKa = 11.84VGRR90 pKa = 11.84YY91 pKa = 7.85LKK93 pKa = 10.35PLEE96 pKa = 3.97EE97 pKa = 4.56RR98 pKa = 11.84LFKK101 pKa = 11.1SLGKK105 pKa = 10.59LFGHH109 pKa = 5.69KK110 pKa = 8.79TVLKK114 pKa = 10.0GVNSQRR120 pKa = 11.84SAEE123 pKa = 3.97LLKK126 pKa = 10.69EE127 pKa = 3.6KK128 pKa = 10.41WEE130 pKa = 4.17MYY132 pKa = 9.84RR133 pKa = 11.84NPVAIGLDD141 pKa = 3.21ASRR144 pKa = 11.84FDD146 pKa = 3.34QHH148 pKa = 8.13VSIDD152 pKa = 3.5ALRR155 pKa = 11.84WEE157 pKa = 4.0HH158 pKa = 7.12SIYY161 pKa = 10.83LEE163 pKa = 4.16CFPQRR168 pKa = 11.84KK169 pKa = 7.59HH170 pKa = 5.33QEE172 pKa = 3.57RR173 pKa = 11.84LRR175 pKa = 11.84RR176 pKa = 11.84LLKK179 pKa = 10.44LQEE182 pKa = 4.56LNSCTGYY189 pKa = 9.13TPDD192 pKa = 5.1GKK194 pKa = 10.79LKK196 pKa = 10.81YY197 pKa = 8.99NVKK200 pKa = 7.88GTRR203 pKa = 11.84MSGDD207 pKa = 3.37MNTSLGNCVLMCSMIKK223 pKa = 10.19AYY225 pKa = 10.77LMSKK229 pKa = 10.6GIDD232 pKa = 3.24GQLANNGDD240 pKa = 3.88DD241 pKa = 3.47CVVFMEE247 pKa = 5.83KK248 pKa = 10.31SDD250 pKa = 4.23LLNFSDD256 pKa = 5.87GLYY259 pKa = 10.85DD260 pKa = 3.33WFLALGFNMAIEE272 pKa = 4.19EE273 pKa = 4.26PVFEE277 pKa = 4.23FEE279 pKa = 5.54QIEE282 pKa = 4.56FCQTKK287 pKa = 9.6PVFDD291 pKa = 3.71GEE293 pKa = 4.2YY294 pKa = 9.9WIMCRR299 pKa = 11.84NPHH302 pKa = 4.89TAINKK307 pKa = 9.47DD308 pKa = 3.4SVMLKK313 pKa = 10.24PFSCSSEE320 pKa = 3.6FRR322 pKa = 11.84GWLDD326 pKa = 3.33AVGTGGLAMAGGIPIFQEE344 pKa = 4.83LYY346 pKa = 9.16RR347 pKa = 11.84TYY349 pKa = 10.53QKK351 pKa = 10.62CGQQRR356 pKa = 11.84KK357 pKa = 9.04ISEE360 pKa = 4.62DD361 pKa = 3.43LLPWSFTNLSKK372 pKa = 11.06GMTRR376 pKa = 11.84QYY378 pKa = 10.57GTVSPAARR386 pKa = 11.84ASFYY390 pKa = 10.01WAFDD394 pKa = 3.26ITPDD398 pKa = 3.56EE399 pKa = 4.4QIALEE404 pKa = 4.78DD405 pKa = 4.47YY406 pKa = 10.67YY407 pKa = 11.63SSTKK411 pKa = 9.97IVSSLGPYY419 pKa = 8.85EE420 pKa = 3.91PRR422 pKa = 11.84GVFAEE427 pKa = 4.2
Molecular weight: 48.85 kDa
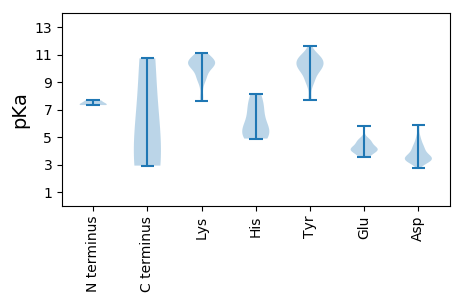
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGN2|A0A1L3KGN2_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 6 OX=1923293 PE=4 SV=1
MM1 pKa = 7.32AVKK4 pKa = 10.55GEE6 pKa = 3.9KK7 pKa = 9.83RR8 pKa = 11.84VIKK11 pKa = 9.55EE12 pKa = 3.54GKK14 pKa = 9.02VFVEE18 pKa = 3.61QRR20 pKa = 11.84YY21 pKa = 9.19NGKK24 pKa = 9.2RR25 pKa = 11.84WVSEE29 pKa = 4.15SSWVSGQQPSKK40 pKa = 10.96EE41 pKa = 4.04GGLTKK46 pKa = 10.49SVARR50 pKa = 11.84KK51 pKa = 9.31IRR53 pKa = 11.84GGVSRR58 pKa = 11.84AGGFVTAPVSGAMITRR74 pKa = 11.84PTVPRR79 pKa = 11.84FGMRR83 pKa = 11.84GDD85 pKa = 3.52STIVSNSEE93 pKa = 3.6LVLNLTPIALAFGVQSLPLIAAQPAWLSTIADD125 pKa = 4.14NYY127 pKa = 10.54SKK129 pKa = 10.26WRR131 pKa = 11.84WISMRR136 pKa = 11.84LIYY139 pKa = 10.34SPKK142 pKa = 10.34CPTTTSGTVAMCLSYY157 pKa = 11.01DD158 pKa = 4.27RR159 pKa = 11.84NDD161 pKa = 3.66APPGSRR167 pKa = 11.84VQLSQTYY174 pKa = 9.63KK175 pKa = 10.79AINFPPYY182 pKa = 10.38AGYY185 pKa = 10.72DD186 pKa = 3.29GAAILNTDD194 pKa = 3.63VTPTSAIYY202 pKa = 10.77LDD204 pKa = 3.69VDD206 pKa = 3.36VTRR209 pKa = 11.84FDD211 pKa = 4.06KK212 pKa = 10.95NWYY215 pKa = 7.69STIGTVAFAALTAFDD230 pKa = 4.16QNQFCPATVHH240 pKa = 6.29IGSDD244 pKa = 3.44GGPAVAVPPGDD255 pKa = 4.55LFFKK259 pKa = 10.7YY260 pKa = 10.28VIEE263 pKa = 4.85LIEE266 pKa = 4.91PINPTMNVV274 pKa = 2.92
MM1 pKa = 7.32AVKK4 pKa = 10.55GEE6 pKa = 3.9KK7 pKa = 9.83RR8 pKa = 11.84VIKK11 pKa = 9.55EE12 pKa = 3.54GKK14 pKa = 9.02VFVEE18 pKa = 3.61QRR20 pKa = 11.84YY21 pKa = 9.19NGKK24 pKa = 9.2RR25 pKa = 11.84WVSEE29 pKa = 4.15SSWVSGQQPSKK40 pKa = 10.96EE41 pKa = 4.04GGLTKK46 pKa = 10.49SVARR50 pKa = 11.84KK51 pKa = 9.31IRR53 pKa = 11.84GGVSRR58 pKa = 11.84AGGFVTAPVSGAMITRR74 pKa = 11.84PTVPRR79 pKa = 11.84FGMRR83 pKa = 11.84GDD85 pKa = 3.52STIVSNSEE93 pKa = 3.6LVLNLTPIALAFGVQSLPLIAAQPAWLSTIADD125 pKa = 4.14NYY127 pKa = 10.54SKK129 pKa = 10.26WRR131 pKa = 11.84WISMRR136 pKa = 11.84LIYY139 pKa = 10.34SPKK142 pKa = 10.34CPTTTSGTVAMCLSYY157 pKa = 11.01DD158 pKa = 4.27RR159 pKa = 11.84NDD161 pKa = 3.66APPGSRR167 pKa = 11.84VQLSQTYY174 pKa = 9.63KK175 pKa = 10.79AINFPPYY182 pKa = 10.38AGYY185 pKa = 10.72DD186 pKa = 3.29GAAILNTDD194 pKa = 3.63VTPTSAIYY202 pKa = 10.77LDD204 pKa = 3.69VDD206 pKa = 3.36VTRR209 pKa = 11.84FDD211 pKa = 4.06KK212 pKa = 10.95NWYY215 pKa = 7.69STIGTVAFAALTAFDD230 pKa = 4.16QNQFCPATVHH240 pKa = 6.29IGSDD244 pKa = 3.44GGPAVAVPPGDD255 pKa = 4.55LFFKK259 pKa = 10.7YY260 pKa = 10.28VIEE263 pKa = 4.85LIEE266 pKa = 4.91PINPTMNVV274 pKa = 2.92
Molecular weight: 29.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
902 |
201 |
427 |
300.7 |
33.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.874 ± 1.112 | 1.663 ± 0.285 |
5.432 ± 0.427 | 5.322 ± 0.998 |
5.1 ± 0.445 | 6.763 ± 0.846 |
0.998 ± 0.395 | 4.878 ± 0.544 |
7.095 ± 1.042 | 8.204 ± 1.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.993 ± 0.387 | 3.991 ± 0.01 |
5.322 ± 0.811 | 3.437 ± 0.063 |
5.765 ± 0.27 | 7.206 ± 1.067 |
5.987 ± 0.838 | 7.317 ± 1.335 |
1.885 ± 0.156 | 3.769 ± 0.136 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
