
Nonlabens ponticola
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Nonlabens
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
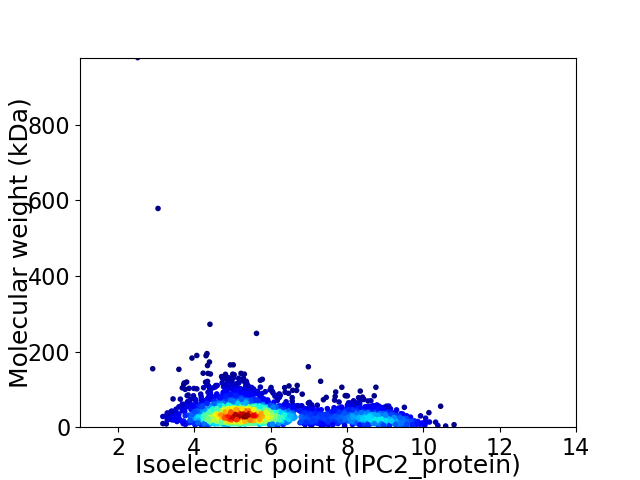
Virtual 2D-PAGE plot for 2545 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S9N180|A0A3S9N180_9FLAO Uncharacterized protein OS=Nonlabens ponticola OX=2496866 GN=EJ995_01345 PE=4 SV=1
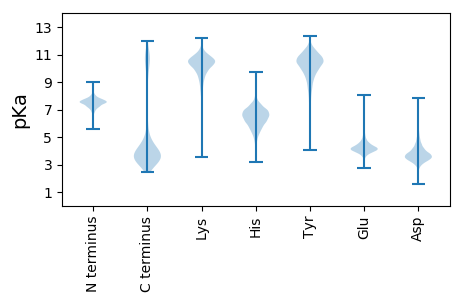
MM1 pKa = 7.32FNKK4 pKa = 10.09IGWFLVIAVTILGCQDD20 pKa = 4.22DD21 pKa = 4.23DD22 pKa = 4.39TEE24 pKa = 4.65FGPVVAPSDD33 pKa = 3.52LSVSFEE39 pKa = 4.14VQGEE43 pKa = 3.99DD44 pKa = 4.8DD45 pKa = 4.48EE46 pKa = 5.24NPNGDD51 pKa = 3.23GTGVVTFSANASNALNYY68 pKa = 8.92TYY70 pKa = 11.18SFGDD74 pKa = 3.42SRR76 pKa = 11.84SSFSSDD82 pKa = 2.3GDD84 pKa = 3.68LEE86 pKa = 4.17HH87 pKa = 7.56RR88 pKa = 11.84YY89 pKa = 8.85VQQGVNTYY97 pKa = 8.15TVTVTATGTGGAATSQTTQVTVFSAFEE124 pKa = 4.69DD125 pKa = 3.6NDD127 pKa = 3.63AKK129 pKa = 10.96QLLTGGSSKK138 pKa = 9.64IWYY141 pKa = 6.48WAAAEE146 pKa = 4.35PGHH149 pKa = 7.39LGVGPNNGDD158 pKa = 3.39PTDD161 pKa = 3.93GNNNFPQFYY170 pKa = 9.58QAAPFEE176 pKa = 4.75KK177 pKa = 10.62NGADD181 pKa = 3.46EE182 pKa = 4.55SLCLYY187 pKa = 10.18EE188 pKa = 4.58DD189 pKa = 3.42QLVFSLNDD197 pKa = 3.64DD198 pKa = 3.88EE199 pKa = 4.94EE200 pKa = 5.0LVYY203 pKa = 10.71QLNNFGQTYY212 pKa = 10.01FNGAFEE218 pKa = 4.56SVVGGSNGFDD228 pKa = 3.1FCYY231 pKa = 10.81DD232 pKa = 3.58FDD234 pKa = 4.37TDD236 pKa = 3.65GVKK239 pKa = 10.44NVTLSPSEE247 pKa = 4.5SIVPDD252 pKa = 3.38SEE254 pKa = 4.57KK255 pKa = 10.77RR256 pKa = 11.84GTTMTFSDD264 pKa = 4.83DD265 pKa = 3.73GFMSYY270 pKa = 10.84YY271 pKa = 10.13INTSVYY277 pKa = 10.49DD278 pKa = 3.62ILEE281 pKa = 4.18LTEE284 pKa = 3.53NRR286 pKa = 11.84MVVRR290 pKa = 11.84GIMNGDD296 pKa = 3.81LAWYY300 pKa = 7.72HH301 pKa = 5.76TFSTDD306 pKa = 4.79DD307 pKa = 3.73PNAEE311 pKa = 4.14QGGDD315 pKa = 3.81DD316 pKa = 3.9NSDD319 pKa = 3.12GSGEE323 pKa = 4.1PEE325 pKa = 4.11GTLVWSDD332 pKa = 3.49EE333 pKa = 4.06FDD335 pKa = 3.45VDD337 pKa = 4.96GAPNPANWTFDD348 pKa = 3.14TGTGSDD354 pKa = 2.8GWGNNEE360 pKa = 3.67QQFYY364 pKa = 10.13TDD366 pKa = 2.96RR367 pKa = 11.84TEE369 pKa = 4.39NIEE372 pKa = 4.08VSNGTLKK379 pKa = 9.02ITARR383 pKa = 11.84DD384 pKa = 3.27EE385 pKa = 4.66DD386 pKa = 3.99FGGRR390 pKa = 11.84SYY392 pKa = 11.03TSSRR396 pKa = 11.84IKK398 pKa = 10.19TEE400 pKa = 3.88GLFEE404 pKa = 4.04FQYY407 pKa = 10.23GTIEE411 pKa = 4.31FSAKK415 pKa = 10.1LPEE418 pKa = 5.59GGGTWPAIWSLGADD432 pKa = 3.74YY433 pKa = 8.5QTNPWPAAGEE443 pKa = 3.85IDD445 pKa = 4.04YY446 pKa = 10.21MEE448 pKa = 6.07HH449 pKa = 6.99IGNTQNTIYY458 pKa = 10.05ATVHH462 pKa = 6.01RR463 pKa = 11.84PGASGGNADD472 pKa = 3.79GGEE475 pKa = 4.04IEE477 pKa = 4.78IEE479 pKa = 4.01NASSEE484 pKa = 4.01FHH486 pKa = 6.53TYY488 pKa = 7.95RR489 pKa = 11.84TIWNEE494 pKa = 3.27DD495 pKa = 3.71TIRR498 pKa = 11.84FFVDD502 pKa = 3.74GEE504 pKa = 4.71LIHH507 pKa = 6.68QVANSDD513 pKa = 3.35AVPFNKK519 pKa = 10.18DD520 pKa = 2.7FFIIMNIAMGGNFGGDD536 pKa = 2.91IDD538 pKa = 4.13PAFTEE543 pKa = 4.22STMEE547 pKa = 3.25VDD549 pKa = 4.35YY550 pKa = 11.35IRR552 pKa = 11.84VYY554 pKa = 10.14QQ555 pKa = 3.67
MM1 pKa = 7.32FNKK4 pKa = 10.09IGWFLVIAVTILGCQDD20 pKa = 4.22DD21 pKa = 4.23DD22 pKa = 4.39TEE24 pKa = 4.65FGPVVAPSDD33 pKa = 3.52LSVSFEE39 pKa = 4.14VQGEE43 pKa = 3.99DD44 pKa = 4.8DD45 pKa = 4.48EE46 pKa = 5.24NPNGDD51 pKa = 3.23GTGVVTFSANASNALNYY68 pKa = 8.92TYY70 pKa = 11.18SFGDD74 pKa = 3.42SRR76 pKa = 11.84SSFSSDD82 pKa = 2.3GDD84 pKa = 3.68LEE86 pKa = 4.17HH87 pKa = 7.56RR88 pKa = 11.84YY89 pKa = 8.85VQQGVNTYY97 pKa = 8.15TVTVTATGTGGAATSQTTQVTVFSAFEE124 pKa = 4.69DD125 pKa = 3.6NDD127 pKa = 3.63AKK129 pKa = 10.96QLLTGGSSKK138 pKa = 9.64IWYY141 pKa = 6.48WAAAEE146 pKa = 4.35PGHH149 pKa = 7.39LGVGPNNGDD158 pKa = 3.39PTDD161 pKa = 3.93GNNNFPQFYY170 pKa = 9.58QAAPFEE176 pKa = 4.75KK177 pKa = 10.62NGADD181 pKa = 3.46EE182 pKa = 4.55SLCLYY187 pKa = 10.18EE188 pKa = 4.58DD189 pKa = 3.42QLVFSLNDD197 pKa = 3.64DD198 pKa = 3.88EE199 pKa = 4.94EE200 pKa = 5.0LVYY203 pKa = 10.71QLNNFGQTYY212 pKa = 10.01FNGAFEE218 pKa = 4.56SVVGGSNGFDD228 pKa = 3.1FCYY231 pKa = 10.81DD232 pKa = 3.58FDD234 pKa = 4.37TDD236 pKa = 3.65GVKK239 pKa = 10.44NVTLSPSEE247 pKa = 4.5SIVPDD252 pKa = 3.38SEE254 pKa = 4.57KK255 pKa = 10.77RR256 pKa = 11.84GTTMTFSDD264 pKa = 4.83DD265 pKa = 3.73GFMSYY270 pKa = 10.84YY271 pKa = 10.13INTSVYY277 pKa = 10.49DD278 pKa = 3.62ILEE281 pKa = 4.18LTEE284 pKa = 3.53NRR286 pKa = 11.84MVVRR290 pKa = 11.84GIMNGDD296 pKa = 3.81LAWYY300 pKa = 7.72HH301 pKa = 5.76TFSTDD306 pKa = 4.79DD307 pKa = 3.73PNAEE311 pKa = 4.14QGGDD315 pKa = 3.81DD316 pKa = 3.9NSDD319 pKa = 3.12GSGEE323 pKa = 4.1PEE325 pKa = 4.11GTLVWSDD332 pKa = 3.49EE333 pKa = 4.06FDD335 pKa = 3.45VDD337 pKa = 4.96GAPNPANWTFDD348 pKa = 3.14TGTGSDD354 pKa = 2.8GWGNNEE360 pKa = 3.67QQFYY364 pKa = 10.13TDD366 pKa = 2.96RR367 pKa = 11.84TEE369 pKa = 4.39NIEE372 pKa = 4.08VSNGTLKK379 pKa = 9.02ITARR383 pKa = 11.84DD384 pKa = 3.27EE385 pKa = 4.66DD386 pKa = 3.99FGGRR390 pKa = 11.84SYY392 pKa = 11.03TSSRR396 pKa = 11.84IKK398 pKa = 10.19TEE400 pKa = 3.88GLFEE404 pKa = 4.04FQYY407 pKa = 10.23GTIEE411 pKa = 4.31FSAKK415 pKa = 10.1LPEE418 pKa = 5.59GGGTWPAIWSLGADD432 pKa = 3.74YY433 pKa = 8.5QTNPWPAAGEE443 pKa = 3.85IDD445 pKa = 4.04YY446 pKa = 10.21MEE448 pKa = 6.07HH449 pKa = 6.99IGNTQNTIYY458 pKa = 10.05ATVHH462 pKa = 6.01RR463 pKa = 11.84PGASGGNADD472 pKa = 3.79GGEE475 pKa = 4.04IEE477 pKa = 4.78IEE479 pKa = 4.01NASSEE484 pKa = 4.01FHH486 pKa = 6.53TYY488 pKa = 7.95RR489 pKa = 11.84TIWNEE494 pKa = 3.27DD495 pKa = 3.71TIRR498 pKa = 11.84FFVDD502 pKa = 3.74GEE504 pKa = 4.71LIHH507 pKa = 6.68QVANSDD513 pKa = 3.35AVPFNKK519 pKa = 10.18DD520 pKa = 2.7FFIIMNIAMGGNFGGDD536 pKa = 2.91IDD538 pKa = 4.13PAFTEE543 pKa = 4.22STMEE547 pKa = 3.25VDD549 pKa = 4.35YY550 pKa = 11.35IRR552 pKa = 11.84VYY554 pKa = 10.14QQ555 pKa = 3.67
Molecular weight: 60.66 kDa
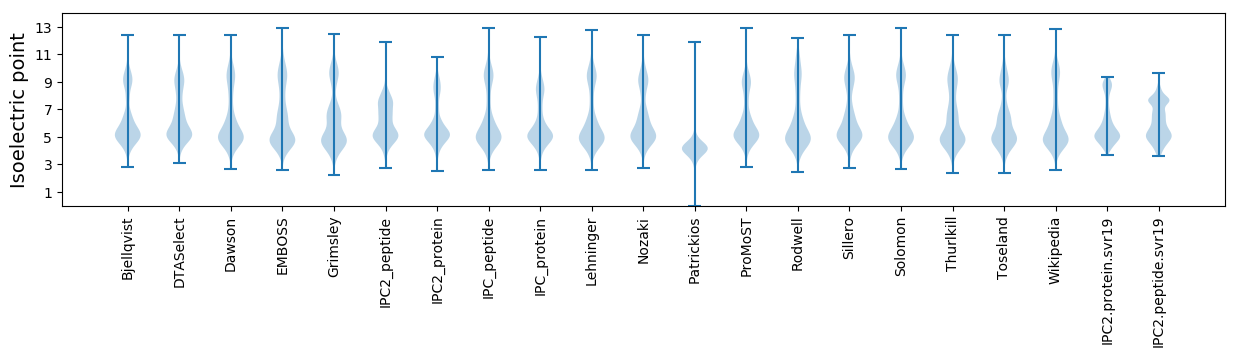
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S9MZT2|A0A3S9MZT2_9FLAO NAD-dependent epimerase/dehydratase family protein OS=Nonlabens ponticola OX=2496866 GN=EJ995_10485 PE=4 SV=1
MM1 pKa = 6.94AQKK4 pKa = 9.31RR5 pKa = 11.84TYY7 pKa = 10.22QPSKK11 pKa = 9.01RR12 pKa = 11.84KK13 pKa = 9.47RR14 pKa = 11.84RR15 pKa = 11.84NKK17 pKa = 9.49HH18 pKa = 3.94GFRR21 pKa = 11.84EE22 pKa = 4.27RR23 pKa = 11.84MASANGRR30 pKa = 11.84KK31 pKa = 9.04VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.09GRR41 pKa = 11.84KK42 pKa = 8.0KK43 pKa = 10.66LSVSTEE49 pKa = 3.69RR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 6.27KK53 pKa = 10.62RR54 pKa = 3.14
MM1 pKa = 6.94AQKK4 pKa = 9.31RR5 pKa = 11.84TYY7 pKa = 10.22QPSKK11 pKa = 9.01RR12 pKa = 11.84KK13 pKa = 9.47RR14 pKa = 11.84RR15 pKa = 11.84NKK17 pKa = 9.49HH18 pKa = 3.94GFRR21 pKa = 11.84EE22 pKa = 4.27RR23 pKa = 11.84MASANGRR30 pKa = 11.84KK31 pKa = 9.04VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.09GRR41 pKa = 11.84KK42 pKa = 8.0KK43 pKa = 10.66LSVSTEE49 pKa = 3.69RR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 6.27KK53 pKa = 10.62RR54 pKa = 3.14
Molecular weight: 6.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
867875 |
29 |
9254 |
341.0 |
38.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.08 ± 0.054 | 0.658 ± 0.015 |
6.929 ± 0.206 | 6.393 ± 0.051 |
4.703 ± 0.039 | 6.557 ± 0.071 |
1.838 ± 0.034 | 7.337 ± 0.045 |
6.302 ± 0.088 | 9.164 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.348 ± 0.039 | 5.507 ± 0.063 |
3.437 ± 0.032 | 3.972 ± 0.035 |
4.145 ± 0.05 | 6.372 ± 0.043 |
5.807 ± 0.062 | 6.469 ± 0.033 |
0.996 ± 0.02 | 3.989 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
